Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03673310
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
23U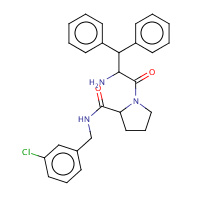 | beta-phenyl-D-phenylalanyl-N-(3- chlorobenzyl)-L-prolinamide | H,I | 3DHK | 0.7 |  |
MPQ | N-METHYL-ALPHA-PHENYL-GLYCINE | D | 1D6E | 0.72 |  |
TFQ | 4-(2,2,2-TRIFLUOROETHYL)-L-PHENYLALANINE | A | 3D6V | 0.71 |  |
FRF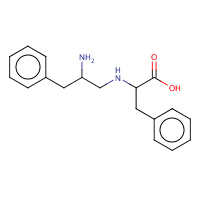 | PHE-REDUCED-PHE | A,I | 1GVX | 0.74 |  |
FCL | 3-CHLORO-L-PHENYLALANINE | E,F | 1OKW | 0.74 |  |
BSI | 2-(BIPHENYL-4-SULFONYL)-1,2,3,4- TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID | A | 1BZS | 0.72 |  |
BSI | 2-(BIPHENYL-4-SULFONYL)-1,2,3,4- TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID | A | 1I76 | 0.72 |  |
153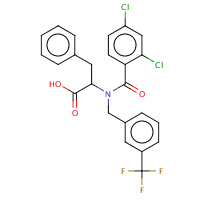 | (2S)-2-[(2,4-DICHLORO-BENZOYL)- (3-TRIFLUOROMETHYL-BENZYL)-AMINO]- 3-PHENYL-PROPIONIC ACID | A,B | 1NHU | 0.85 |  |
ZAE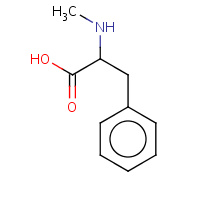 | N-methyl-D-phenylalanine | H,I,R | 1TBZ | 0.7 |  |
200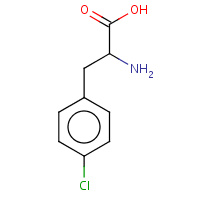 | 4-CHLORO-L-PHENYLALANINE | A,B | 2AKW | 0.75 |  |
IAM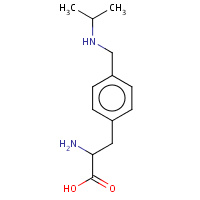 | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XY5 | 0.73 |  |
IAM | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XY8 | 0.73 |  |
IAM | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XY9 | 0.73 |  |
IAM | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XXZ | 0.73 |  |
IAM | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XY4 | 0.73 |  |
IAM | 4-[(ISOPROPYLAMINO)METHYL]PHENYLALANINE | A | 1XY6 | 0.73 |  |
MEA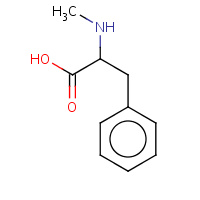 | N-METHYLPHENYLALANINE | A | 2PIL | 0.7 |  |
MEA | N-METHYLPHENYLALANINE | A,B,C,D | 1H0I | 0.7 |  |
MEA | N-METHYLPHENYLALANINE | A,B,I | 1DOJ | 0.7 |  |
22U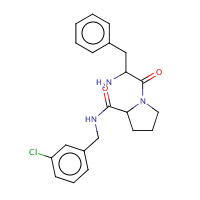 | D-phenylalanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZC9 | 0.71 |  |
P21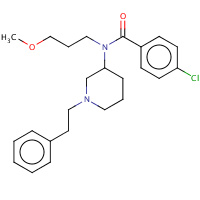 | 4-chloro-N-(3-methoxypropyl)-N- [(3S)-1-(2-phenylethyl)piperidin- 3-yl]benzamide | A | 2VD4 | 0.72 |  |


