Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03128788
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
RNS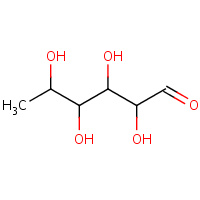 | L-RHAMNOSE | A,B,C,D | 1X8D | 0.75 |  |
RNS | L-RHAMNOSE | A,B,C,D | 1DE6 | 0.75 |  |
RNS | L-RHAMNOSE | A,B,C,D | 2I56 | 0.75 |  |
PAN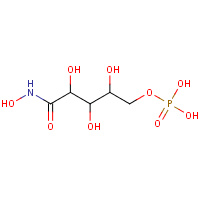 | 5-PHOSPHO-D-ARABINOHYDROXAMIC ACID | A,B | 1KOJ | 0.78 |  |
PAN | 5-PHOSPHO-D-ARABINOHYDROXAMIC ACID | A,B | 2GC0 | 0.78 |  |
XYH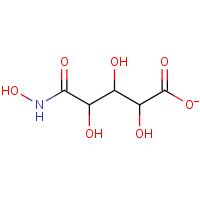 | XYLAROHYDROXAMATE | A,B,C,D | 1EC9 | 0.8 |  |
LLH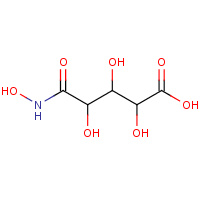 | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)- 5-OXOPENTANOIC ACID | A,B,C,D,E,F | 2PP1 | 0.8 |  |
GLG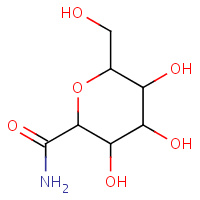 | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE | A | 1GG8 | 0.79 |  |
GLO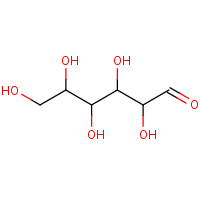 | D-GLUCOSE IN LINEAR FORM | A,B | 1XYB | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FQD | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A,B | 1XYM | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A,B | 1EZ9 | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FBO | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1AC0 | 0.72 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FQC | 0.72 |  |
CBF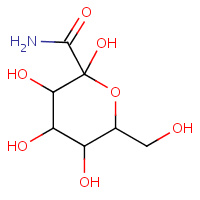 | C-(1-HYDROGYL-BETA-D-GLUCOPYRANOSYL) FORMAMIDE | A | 1P4J | 0.74 |  |
DNO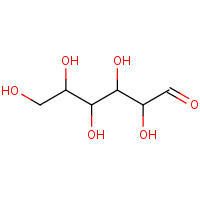 | D-mannose | A | 3BDK | 0.72 |  |
XLS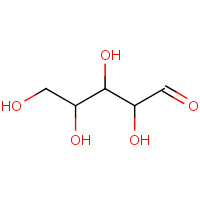 | D-XYLOSE (LINEAR FORM) | A,B | 2QW5 | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 2BRP | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 3XIS | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 8XIM | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 4XIS | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 9XIM | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 5XIN | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 8XIA | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 6XIM | 0.71 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 1XIC | 0.71 |  |
AOS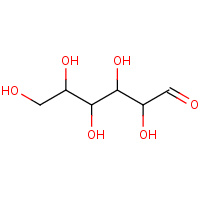 | D-ALLOSE | A,B,C,D | 2I57 | 0.72 |  |
EHM | (2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | A | 2HXT | 0.75 |  |


