Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02518927
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
C2A | 1-(3-CHLOROPHENYL)METHANAMINE | B,I | 2C8Z | 0.71 |  |
LY1 | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO- 1H-BENZO[C]AZEPINE | A,B | 1N7I | 0.79 |  |
19U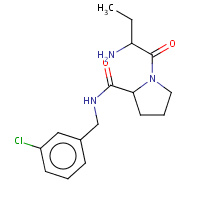 | 1-[(2R)-2-aminobutanoyl]-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZFP | 0.76 |  |
SRE | (1S,4S)-4-(3,4-dichlorophenyl)- N-methyl-1,2,3,4-tetrahydronaphthalen- 1-amine | A | 3GWU | 0.8 |  |
GVQ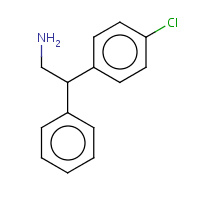 | (2R)-2-(4-CHLOROPHENYL)-2-PHENYLETHANAMINE | A | 2UW8 | 0.75 |  |
22U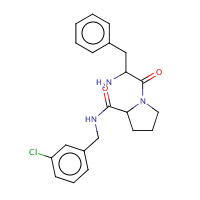 | D-phenylalanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZC9 | 0.78 |  |
CPU | A,B | 1CR6 | 0.7 |  | |
JNH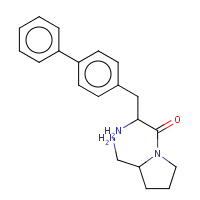 | 1-[2-(S)-AMINO-3-BIPHENYL-4-YL- PROPIONYL]-PYRROLIDINE-2-(S)-CARBONITRILE | I,J | 2AJL | 0.71 |  |
16U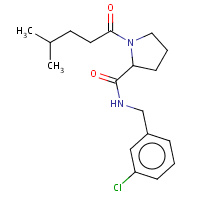 | N-(3-chlorobenzyl)-1-(4-methylpentanoyl)- L-prolinamide | H,I | 3DT0 | 0.76 |  |
SKA | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE | A,B | 1YZ3 | 0.78 |  |
176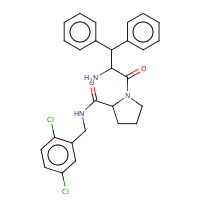 | 1-(2-AMINO-3,3-DIPHENYL-PROPIONYL)- PYRROLIDINE-3-CARBOXYLIC ACID 2,5- DICHLORO-BENZYLAMIDE | A,B | 1TA2 | 0.73 |  |
C2B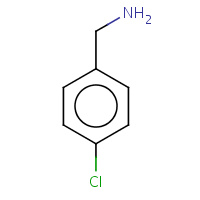 | 1-(4-CHLOROPHENYL)METHANAMINE | D,H | 2Q7Q | 0.71 |  |
V35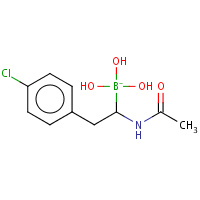 | D-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID | B,C | 2VGC | 0.73 |  |
23U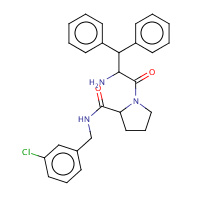 | beta-phenyl-D-phenylalanyl-N-(3- chlorobenzyl)-L-prolinamide | H,I | 3DHK | 0.74 |  |
21U | D-leucyl-N-(3-chlorobenzyl)-L-prolinamide | H,I | 2ZGB | 0.76 |  |
V36 | L-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID | B,C | 1VGC | 0.73 |  |
64U | 3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 3DUX | 0.78 |  |
QN3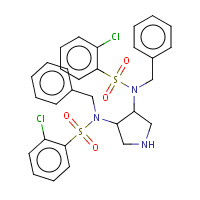 | N,N'-(3S,4S)-pyrrolidine-3,4-diylbis(N- benzyl-2-chlorobenzenesulfonamide) | A,B | 2QNQ | 0.72 |  |


