Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02459077
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
GCN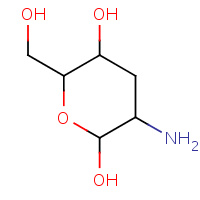 | 3-DEOXY-D-GLUCOSAMINE | A | 1FI1 | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFG | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QKC | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A,B | 2GRX | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QJQ | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFF | 0.71 |  |
LYZ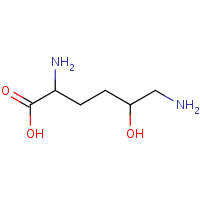 | 5-HYDROXYLYSINE | A,B,C,D | 1QGW | 0.75 |  |
LYZ | 5-HYDROXYLYSINE | A,B,C,D | 1XG0 | 0.75 |  |
LYZ | 5-HYDROXYLYSINE | A,B,C,D | 1XF6 | 0.75 |  |
LYZ | 5-HYDROXYLYSINE | A,B,C | 1YGV | 0.75 |  |
LYZ | 5-HYDROXYLYSINE | A,B,C | 1Y0F | 0.75 |  |
GDL | 2-ACETAMIDO-2-DEOXY-D-GLUCONO-1,5- LACTONE | A,B | 1UR9 | 0.77 |  |
GDL | 2-ACETAMIDO-2-DEOXY-D-GLUCONO-1,5- LACTONE | A,B,C,D,E,F | 1O7A | 0.77 |  |
GDL | 2-ACETAMIDO-2-DEOXY-D-GLUCONO-1,5- LACTONE | A,B | 1UR8 | 0.77 |  |
EKE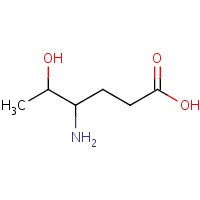 | (4S,5R)-4-AMINO-5-HYDROXYHEXANOIC ACID | A,I | 2HAL | 0.71 |  |
1GN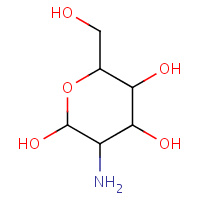 | 2-DEOXY-2-AMINOGALACTOSE | A,B | 3GAL | 0.71 |  |
W72 | 6-DEOXY-6-[(2R,3R,4R)-3,4-DIHYDROXY- 2-(HYDROXYMETHYL)PYRROLIDIN-1-YL]- L-GULONIC ACID | A | 2FYV | 0.73 |  |
CBF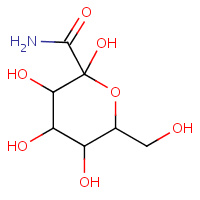 | C-(1-HYDROGYL-BETA-D-GLUCOPYRANOSYL) FORMAMIDE | A | 1P4J | 0.71 |  |
AR4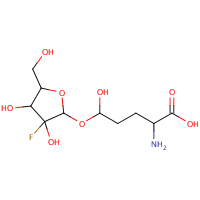 | 2-AMINO-5-(3-FLUORO-3,4-DIHYDROXY- 5-HYDROXYMETHYL-TETRAHYDRO-FURAN- 2-YLOXY)-5-HYDROXY-PENTANOIC ACID | A,B,C | 1S2D | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1PBR | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QKC | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFG | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1FI1 | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A,B,C,D | 3FXI | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFF | 0.71 |  |
KAH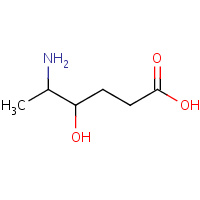 | 5-AMINO-4-HYDROXYHEXANOIC ACID | A | 1H7P | 0.7 |  |
GCS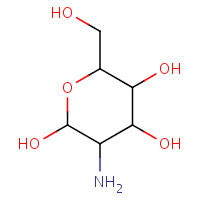 | D-GLUCOSAMINE | A | 3CO4 | 0.71 |  |
GCS | D-GLUCOSAMINE | A | 1QGI | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZS | 0.71 |  |
GCS | D-GLUCOSAMINE | A | 1E9L | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B,C,D | 3FXI | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZV | 0.71 |  |
GLG | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE | A | 1GG8 | 0.73 |  |
NDZ | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino- 2-carboxyethyl]-6,7-dihydroxyhexahydro- 2H-furo[3,2-b]pyran-2-carboxylic acid | A | 2ZNU | 0.72 |  |
MS8 | (2R,3aR,7aR)-2-[(2S)-2-amino-3- hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a- hexahydrofuro[4,5-b]pyran-2-carboxylic acid | A,B | 3GBB | 0.73 |  |
ILX | 4,5-DIHYDROXYISOLEUCINE | A,B,C,I,J,L,M | 3CQZ | 0.71 |  |
ILX | 4,5-DIHYDROXYISOLEUCINE | A,B,C,I,J,L,M | 1K83 | 0.71 |  |
ILX | 4,5-DIHYDROXYISOLEUCINE | A,B,C,I,J,L, M,T | 2VUM | 0.71 |  |
MUR | MURAMIC ACID | A,C,E,G | 1LOD | 0.72 |  |


