Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02183317
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
MYG | GLUCOSAMINYL-(ALPHA-6)-D-MYO-INOSITOL | A | 1GYM | 0.76 |  |
B19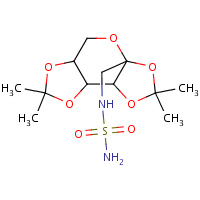 | N-{[(3AS,5AR,8AR,8BS)-2,2,7,7-TETRAMETHYLTETRAHYDRO- 3AH-BIS[1,3]DIOXOLO[4,5-B:4',5'- D]PYRAN-3A-YL]METHYL}SULFAMIDE | A | 2H15 | 0.9 |  |
AD7 | octyl 3-amino-3-deoxy-2-O-(2,6- dideoxy-alpha-L-lyxo-hexopyranosyl)- beta-D-galactopyranoside | A | 2RJ9 | 0.71 |  |
AD7 | octyl 3-amino-3-deoxy-2-O-(2,6- dideoxy-alpha-L-lyxo-hexopyranosyl)- beta-D-galactopyranoside | A | 2RJ4 | 0.71 |  |
TOA | 3-DEOXY-3-AMINO GLUCOSE | A | 1TOB | 0.7 |  |
TOA | 3-DEOXY-3-AMINO GLUCOSE | A | 2TOB | 0.7 |  |
N30 | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2- [(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]- 3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY- ALPHA-D-GLUCOPYRANOSIDE | B | 2O3X | 0.74 |  |
GCS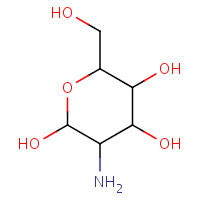 | D-GLUCOSAMINE | A | 3CO4 | 0.74 |  |
GCS | D-GLUCOSAMINE | A | 1QGI | 0.74 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZS | 0.74 |  |
GCS | D-GLUCOSAMINE | A | 1E9L | 0.74 |  |
GCS | D-GLUCOSAMINE | A,B,C,D | 3FXI | 0.74 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZV | 0.74 |  |
NMY | NEOMYCIN | A | 2B0Q | 0.72 |  |
NMY | NEOMYCIN | 4,B | 2QP1 | 0.72 |  |
NMY | NEOMYCIN | A,B | 2ET4 | 0.72 |  |
NMY | NEOMYCIN | A,E,N | 2QAN | 0.72 |  |
NMY | NEOMYCIN | A,B | 2FCY | 0.72 |  |
NMY | NEOMYCIN | A | 2QP0 | 0.72 |  |
NMY | NEOMYCIN | A | 1I9V | 0.72 |  |
NMY | NEOMYCIN | A | 2QAL | 0.72 |  |
NMY | NEOMYCIN | 4,B | 2QOZ | 0.72 |  |
NMY | NEOMYCIN | A,C | 2A04 | 0.72 |  |
NMY | NEOMYCIN | A | 1EI2 | 0.72 |  |
NMY | NEOMYCIN | A,B | 3C7R | 0.72 |  |
NMY | NEOMYCIN | 4,B | 2QAM | 0.72 |  |
NMY | NEOMYCIN | 4,B | 2QAO | 0.72 |  |
NMY | NEOMYCIN | A | 2QOY | 0.72 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1PBR | 0.74 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QKC | 0.74 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFG | 0.74 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1FI1 | 0.74 |  |
PA1 | PAROMOMYCIN (RING 1) | A,B,C,D | 3FXI | 0.74 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFF | 0.74 |  |
BDG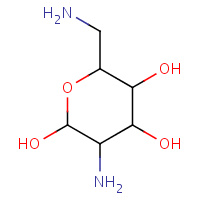 | O-2,6-DIAMINO-2,6-DIDEOXY-ALPHA- D-GLUCOPYRANOSE | A | 1QD3 | 0.72 |  |
BDG | O-2,6-DIAMINO-2,6-DIDEOXY-ALPHA- D-GLUCOPYRANOSE | B | 1O9M | 0.72 |  |
BDG | O-2,6-DIAMINO-2,6-DIDEOXY-ALPHA- D-GLUCOPYRANOSE | A | 1NEM | 0.72 |  |
IDG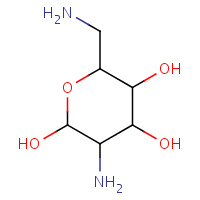 | O-2,6-DIAMINO-2,6-DIDEOXY-BETA- L-IDOPYRANOSE | A | 1QD3 | 0.72 |  |
IDG | O-2,6-DIAMINO-2,6-DIDEOXY-BETA- L-IDOPYRANOSE | A | 1NEM | 0.72 |  |
IDG | O-2,6-DIAMINO-2,6-DIDEOXY-BETA- L-IDOPYRANOSE | A | 1PBR | 0.72 |  |
NGO | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY- ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3- OXAZOLE | A,B,C,D | 1E6Z | 0.72 |  |
1GN | 2-DEOXY-2-AMINOGALACTOSE | A,B | 3GAL | 0.74 |  |
AIG | 4-AMINO-2-HEXYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R7Y | 0.77 |  |
AIG | 4-AMINO-2-HEXYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R7X | 0.77 |  |
AIG | 4-AMINO-2-HEXYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R81 | 0.77 |  |
AIG | 4-AMINO-2-HEXYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R7V | 0.77 |  |
AIG | 4-AMINO-2-HEXYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R80 | 0.77 |  |
GDA | 4-DEOXY-4-AMINO-BETA-D-GLUCOSE | A,B | 1OCB | 0.72 |  |
AOG | 4-AMINO-2-OCTYLOXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-3,5-DIOL | A | 1R82 | 0.74 |  |
XXX | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)- 4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)- 5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO- 2H-PYRAN-3,4-DIOL | B | 2F4S | 0.74 |  |
XXX | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)- 4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)- 5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO- 2H-PYRAN-3,4-DIOL | A,B | 2FCX | 0.74 |  |
XXX | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)- 4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)- 5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO- 2H-PYRAN-3,4-DIOL | B | 2ET8 | 0.74 |  |
MUR | MURAMIC ACID | A,C,E,G | 1LOD | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1CPU | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1NM9 | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1MFV | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1PIG | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | X | 1Z32 | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1MFU | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 3BLK | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | X | 3BLP | 0.73 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 3DHP | 0.73 |  |
TOC | 2,3,6-TRIDEOXY-2,6-DIAMINO GLUCOSE | A | 2TOB | 0.71 |  |
TOC | 2,3,6-TRIDEOXY-2,6-DIAMINO GLUCOSE | A | 1TOB | 0.71 |  |
RIO | RIBOSTAMYCIN | A,B | 1M4G | 0.74 |  |
RIO | RIBOSTAMYCIN | A,B | 3C3Z | 0.74 |  |
RIO | RIBOSTAMYCIN | A,B | 1S3Z | 0.74 |  |
RIO | RIBOSTAMYCIN | A,B,C,D | 2FCZ | 0.74 |  |
RIO | RIBOSTAMYCIN | A,B | 2ET5 | 0.74 |  |
RIO | RIBOSTAMYCIN | A,B | 3DVV | 0.74 |  |
RIO | RIBOSTAMYCIN | A | 2BUE | 0.74 |  |
DAG | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A | 2PIK | 0.73 |  |
DAG | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A | 6CGT | 0.73 |  |
DAG | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A,B | 1PIK | 0.73 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1FI1 | 0.74 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFG | 0.74 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QKC | 0.74 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A,B | 2GRX | 0.74 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QJQ | 0.74 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFF | 0.74 |  |
PAR | PAROMOMYCIN | A | 1FYP | 0.72 |  |
PAR | PAROMOMYCIN | V,Z | 2J02 | 0.72 |  |
PAR | PAROMOMYCIN | Z | 2UXD | 0.72 |  |
PAR | PAROMOMYCIN | A,D,N,Y,Z | 1N33 | 0.72 |  |
PAR | PAROMOMYCIN | V,Y,Z | 2WDM | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2VQE | 0.72 |  |
PAR | PAROMOMYCIN | A,D,L,N | 1IBK | 0.72 |  |
PAR | PAROMOMYCIN | Z | 2UXB | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2UU9 | 0.72 |  |
PAR | PAROMOMYCIN | Z | 2UXC | 0.72 |  |
PAR | PAROMOMYCIN | A,D,N,X | 1XMQ | 0.72 |  |
PAR | PAROMOMYCIN | B | 2O3W | 0.72 |  |
PAR | PAROMOMYCIN | V,Y,Z | 2WDG | 0.72 |  |
PAR | PAROMOMYCIN | A | 2Z4K | 0.72 |  |
PAR | PAROMOMYCIN | 4,B | 2Z4N | 0.72 |  |
PAR | PAROMOMYCIN | V,Y,Z | 2WDK | 0.72 |  |
PAR | PAROMOMYCIN | A,E | 2Z4M | 0.72 |  |
PAR | PAROMOMYCIN | A,B,C,D | 3BNQ | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2UUB | 0.72 |  |
PAR | PAROMOMYCIN | A,D,H,N | 1FJG | 0.72 |  |
PAR | PAROMOMYCIN | A,B | 1J7T | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2UUA | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2UUC | 0.72 |  |
PAR | PAROMOMYCIN | A,D,J,N,X | 1XNQ | 0.72 |  |
PAR | PAROMOMYCIN | A,D,E,J,M,N,Y | 1N32 | 0.72 |  |
PAR | PAROMOMYCIN | A | 2VQY | 0.72 |  |
PAR | PAROMOMYCIN | A,B | 3C44 | 0.72 |  |
PAR | PAROMOMYCIN | V,Z | 2J00 | 0.72 |  |
PAR | PAROMOMYCIN | 4,B | 2Z4L | 0.72 |  |
PAR | PAROMOMYCIN | V,Y,Z | 2WDH | 0.72 |  |
PAR | PAROMOMYCIN | A,D,J,N,Y | 1IBL | 0.72 |  |
PAR | PAROMOMYCIN | A,B,C,D | 3BNR | 0.72 |  |
PAR | PAROMOMYCIN | A,D,J,N,X | 1XMO | 0.72 |  |
PAR | PAROMOMYCIN | Y,Z | 2VQF | 0.72 |  |
PAR | PAROMOMYCIN | A,D,J,N,X | 1XNR | 0.72 |  |


