Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs01231992
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
LAZ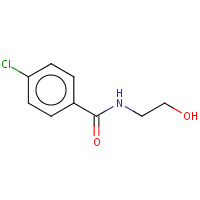 | N-(2-AMINOETHYL)-P-CHLOROBENZAMIDE | A,B | 1OJC | 0.75 |  |
SKA | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE | A,B | 1YZ3 | 0.75 |  |
SP8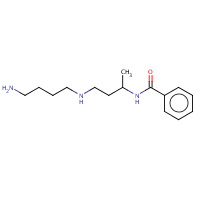 | N-{(1S)-3-[(4-aminobutyl)amino]- 1-methylpropyl}benzamide | A,B | 3CNS | 0.7 |  |
23U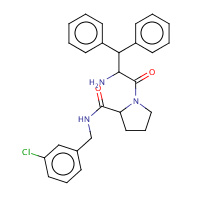 | beta-phenyl-D-phenylalanyl-N-(3- chlorobenzyl)-L-prolinamide | H,I | 3DHK | 0.72 |  |
176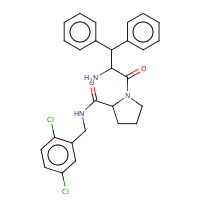 | 1-(2-AMINO-3,3-DIPHENYL-PROPIONYL)- PYRROLIDINE-3-CARBOXYLIC ACID 2,5- DICHLORO-BENZYLAMIDE | A,B | 1TA2 | 0.72 |  |
CRP | ((1RS,3SR)-2,2-DICHLORO-N-[(R)- 1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL- 3-METHYLCYCLOPROPANECARBOXAMIDE | A | 2STD | 0.75 |  |
CRP | ((1RS,3SR)-2,2-DICHLORO-N-[(R)- 1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL- 3-METHYLCYCLOPROPANECARBOXAMIDE | A,B,C | 7STD | 0.75 |  |
21U | D-leucyl-N-(3-chlorobenzyl)-L-prolinamide | H,I | 2ZGB | 0.74 |  |
V35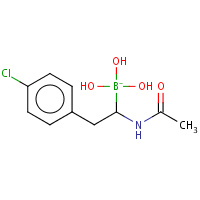 | D-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID | B,C | 2VGC | 0.75 |  |
SRE | (1S,4S)-4-(3,4-dichlorophenyl)- N-methyl-1,2,3,4-tetrahydronaphthalen- 1-amine | A | 3GWU | 0.74 |  |
64U | 3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 3DUX | 0.74 |  |
16U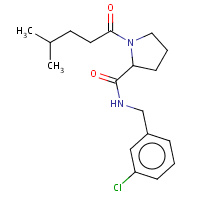 | N-(3-chlorobenzyl)-1-(4-methylpentanoyl)- L-prolinamide | H,I | 3DT0 | 0.74 |  |
22U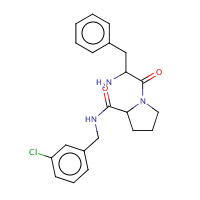 | D-phenylalanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZC9 | 0.76 |  |
2O7 | 7A-[(4-cyanophenyl)methyl]-6-(3,5- dichlorophenyl)-5-oxo-2,3,5,7A- tetrahydro-1H-pyrrolo[1,2-A]pyrrole- 7-carbonitrile | A | 2O7N | 0.74 |  |
V36 | L-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID | B,C | 1VGC | 0.75 |  |
19U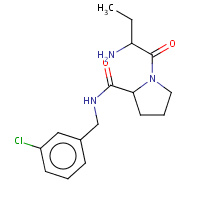 | 1-[(2R)-2-aminobutanoyl]-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZFP | 0.74 |  |
SP9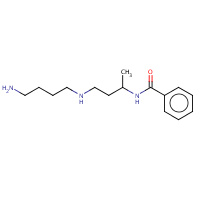 | N-{(1R)-3-[(4-aminobutyl)amino]- 1-methylpropyl}benzamide | A,B | 3CNT | 0.7 |  |
P21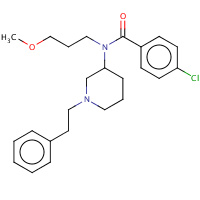 | 4-chloro-N-(3-methoxypropyl)-N- [(3S)-1-(2-phenylethyl)piperidin- 3-yl]benzamide | A | 2VD4 | 0.72 |  |
LY1 | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO- 1H-BENZO[C]AZEPINE | A,B | 1N7I | 0.75 |  |


