Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00803121
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
1SQ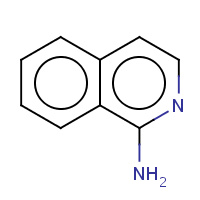 | ISOQUINOLIN-1-AMINE | A | 2OHK | 0.82 |  |
1BY | [2-(PYRIDIN-2-YLAMINO)ETHANE-1,1- DIYL]BIS(PHOSPHONIC ACID) | A,B | 2I19 | 0.83 |  |
11X | N-(pyridin-3-ylmethyl)aniline | A | 3EJ0 | 0.82 |  |
2AP | 2-AMINOPYRIDINE | A | 1AEO | 0.85 |  |
246 | 1-benzyl-3-(2-chloropyridin-4-yl)urea | A | 2QPM | 0.74 |  |
3MP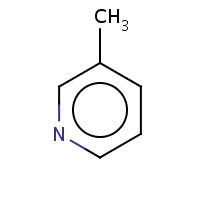 | 3-METHYLPYRIDINE | A | 1EUB | 0.78 |  |
3MP | 3-METHYLPYRIDINE | A | 1BM6 | 0.78 |  |
3AP | 3-AMINOPYRIDINE | A | 1AEF | 0.74 |  |
2AQ | QUINOLIN-2-AMINE | A | 2OHL | 0.73 |  |
218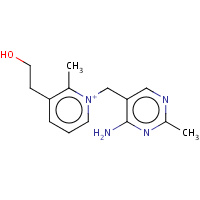 | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5- YL)METHYL]-3-(2-HYDROXYETHYL)-2- METHYLPYRIDINIUM | A | 2HOP | 0.73 |  |
26D | PYRIDINE-2,6-DIAMINE | A | 2ANZ | 0.78 |  |
275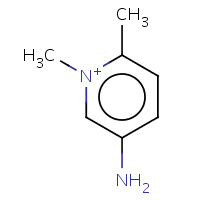 | 5-amino-1,2-dimethylpyridinium | X | 2RBW | 0.71 |  |
5BP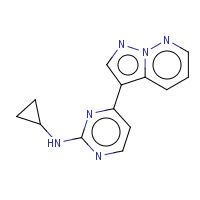 | N-cyclopropyl-4-pyrazolo[1,5-b]pyridazin- 3-ylpyrimidin-2-amine | A,C | 3EJ1 | 0.72 |  |
5MS | N-{2-methyl-5-[(6-phenylpyrimidin- 4-yl)amino]phenyl}methanesulfonamide | A | 3EXO | 0.72 |  |
286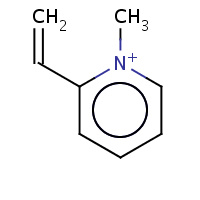 | 2-ethenyl-1-methylpyridinium | X | 2RC2 | 0.72 |  |
245 | 1-(2-chloropyridin-4-yl)-3-phenylurea | A | 2QKN | 0.72 |  |
6IP | 6-[2-(1H-INDOL-6-YL)ETHYL]PYRIDIN- 2-AMINE | A | 2OHP | 0.75 |  |
5SC | 3-((3-bromo-5-o-tolylpyrazolo[1,5- a]pyrimidin-7-ylamino)methyl)pyridine 1- oxide | A | 2R3Q | 0.7 |  |
4AP | 4-AMINOPYRIDINE | A | 1AEG | 0.75 |  |
5IQ | ISOQUINOLIN-5-AMINE | A,B | 2F2T | 0.73 |  |
084 | 4-[5-[2-(1-PHENYL-ETHYLAMINO)-PYRIMIDIN- 4-YL]-1-METHYL-4-(3-TRIFLUOROMETHYLPHENYL)- 1H-IMIDAZOL-2-YL]-PIPERIDINE | A | 1OUK | 0.74 |  |


