Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00717490
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
1AN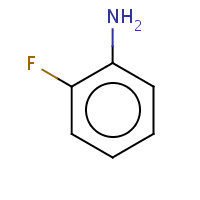 | 2-FLUOROANILINE | A | 1LGW | 0.72 |  |
DSM | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN- 5-YL)-N-METHYLPROPAN-1-AMINE | A | 2QJU | 0.73 |  |
DSM | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN- 5-YL)-N-METHYLPROPAN-1-AMINE | A | 2QB4 | 0.73 |  |
AU4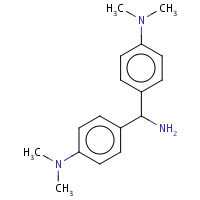 | 4,4'-(AMINOMETHYLENE)BIS(N,N-DIMETHYLANILINE) | A | 2PYZ | 0.73 |  |
URS | N-PHENYLTHIOUREA | A,B | 1BUG | 0.86 |  |
NYL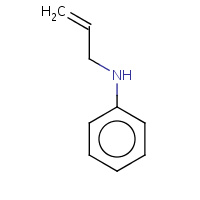 | N-ALLYL-ANILINE | A | 1OVK | 0.81 |  |
264 | (phenylamino)acetonitrile | A | 2RBN | 0.81 |  |
MPX | 4-({[(4-METHYLPIPERAZIN-1-YL)AMINO]CARBONOTHIOYL}AMINO)BENZENESULFONAMIDE | A | 1ZH9 | 0.72 |  |
NP2 | N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | A | 1WUM | 0.72 |  |
PHJ | N-[(AMINOOXY)CARBONYL]-N-PHENYLAMINE | A,B | 1UR9 | 0.7 |  |
IDM | INDOLINE | A,B | 3CEP | 0.7 |  |
IDM | INDOLINE | A | 1AEK | 0.7 |  |
BSU | 1,3-DIPHENYLUREA | A | 3E85 | 0.78 |  |
BSU | 1,3-DIPHENYLUREA | A | 2ZJF | 0.78 |  |
171 | 2-PHENYLAMINO-ETHANESULFONIC ACID | A,B | 1SXG | 0.72 |  |
1MR | N-METHYLANILINE | X | 2OTZ | 0.81 |  |
CIU | N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA | A,B | 1EK1 | 0.8 |  |
CIU | N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA | A | 1VJ5 | 0.8 |  |
PL0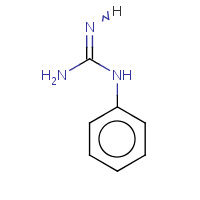 | 1-phenylguanidine | A | 2O8W | 0.78 |  |


