Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00483677
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
MNL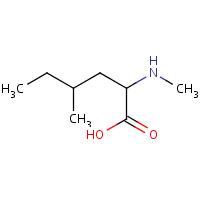 | 4,N-DIMETHYLNORLEUCINE | C | 1CWC | 0.75 |  |
SSC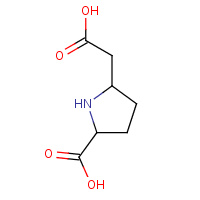 | (2S,5S)-5-CARBOXYMETHYLPROLINE | A,B,C,D | 1Q19 | 0.8 |  |
BG5 | 5-HYDROXYAMINO-3-METHYL-PYRROLIDINE- 2-CARBOXYLIC ACID | A | 1TV2 | 0.7 |  |
CPV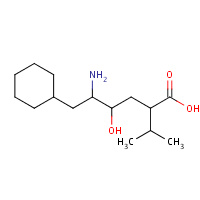 | 5-AMINO-6-CYCLOHEXYL-4-HYDROXY- 2-ISOPROPYL-HEXANOIC ACID | I | 1IVQ | 0.7 |  |
RPH | 6-HYDROXO-OCTAHYDRO-INDOLE-2-CARBALDEHYDE | H,I | 1A2C | 0.83 |  |
LY5 | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy- 4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline- 3-carboxylic acid | A,B,C,D | 2QS4 | 0.74 |  |
MCP | 2-CARBOXY-4-METHYLPIPERIDINE | H | 1ETR | 0.75 |  |
TMD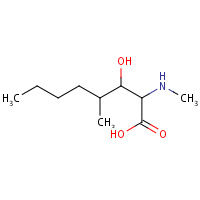 | (6,7-DIHYDRO)4-[(E)-BUTENYL]-4,N- DIMETHYL-THREONINE | C | 1CWK | 0.72 |  |
4FB | (4S)-4-FLUORO-L-PROLINE | A | 2Q6P | 0.71 |  |
POM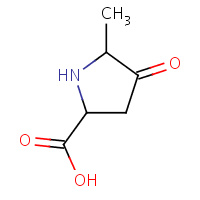 | CIS-5-METHYL-4-OXOPROLINE | A,B | 1A7Z | 0.73 |  |
OIC | OCTAHYDROINDOLE-2-CARBOXYLIC ACID | B,C | 3BV9 | 1 |  |
OIC | OCTAHYDROINDOLE-2-CARBOXYLIC ACID | A | 1BDK | 1 |  |
ABX | 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]- 4-(METHOXYCARBONYL)PROLINE | A | 1XOE | 0.72 |  |
X7O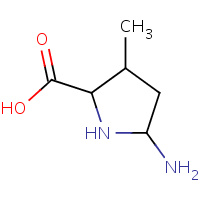 | 5-AMINO-3-METHYL-PYRROLIDINE-2- CARBOXYLIC ACID | A | 1L2Q | 0.73 |  |
NLH | (2R,5S)-5-amino-2-butyl-4,4-dihydroxynonanoic acid | A,B,I | 3DCR | 0.71 |  |
ZAL | 3-cyclohexyl-D-alanine | I | 1HBT | 0.76 |  |
NLK | (2R,5S)-5-amino-2-butyl-4-oxononanoic acid | A,B,I | 3DCK | 0.71 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D | 1D5Z | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | H,I | 5GDS | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | H,P | 2A2X | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D,H | 2FEQ | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | H,P | 2ANK | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A,C,D | 3DPP | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A | 1EB1 | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D,H | 1NZQ | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | I | 1HBT | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D | 1D6E | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A,B | 1B3H | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D,H | 2FES | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | C,D,E,G,H | 3DPQ | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | I | 1THS | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | I | 1QUR | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A,B,C,D | 3DPO | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | H,I | 4THN | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A,D | 1D5M | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D,H | 1O0D | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P | 1YWH | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | A | 1XRZ | 0.76 |  |
ALC | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID | D | 1D5X | 0.76 |  |
CHG | CYCLOHEXYL-GLYCINE | 2,3,4 | 8KME | 0.77 |  |
CHG | CYCLOHEXYL-GLYCINE | H,I | 4THN | 0.77 |  |
CHG | CYCLOHEXYL-GLYCINE | H,I,J | 7KME | 0.77 |  |
CHG | CYCLOHEXYL-GLYCINE | H,I | 5GDS | 0.77 |  |
DPR | D-PROLINE | P | 1MCB | 0.73 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5B | 0.73 |  |
DPR | D-PROLINE | A | 2KDQ | 0.73 |  |
DPR | D-PROLINE | P | 1MCS | 0.73 |  |
DPR | D-PROLINE | A,B | 2AXI | 0.73 |  |
DPR | D-PROLINE | A | 1JY9 | 0.73 |  |
DPR | D-PROLINE | A | 2JUE | 0.73 |  |
DPR | D-PROLINE | P | 1MCK | 0.73 |  |
DPR | D-PROLINE | P | 1MCR | 0.73 |  |
DPR | D-PROLINE | A | 2D55 | 0.73 |  |
DPR | D-PROLINE | A | 1T8J | 0.73 |  |
DPR | D-PROLINE | A,B | 2EJ6 | 0.73 |  |
DPR | D-PROLINE | P | 1MCE | 0.73 |  |
DPR | D-PROLINE | P | 1MCI | 0.73 |  |
DPR | D-PROLINE | A,B | 1SNE | 0.73 |  |
DPR | D-PROLINE | A,B,C | 1QFI | 0.73 |  |
DPR | D-PROLINE | A,P | 1MCD | 0.73 |  |
DPR | D-PROLINE | A | 1ICL | 0.73 |  |
DPR | D-PROLINE | P | 1MCQ | 0.73 |  |
DPR | D-PROLINE | G,M,P,S | 2I5Y | 0.73 |  |
DPR | D-PROLINE | A,B,C,D | 3BOG | 0.73 |  |
DPR | D-PROLINE | A,L | 1ZEA | 0.73 |  |
DPR | D-PROLINE | A,B | 173D | 0.73 |  |
DPR | D-PROLINE | A,P | 1MCJ | 0.73 |  |
DPR | D-PROLINE | A,B,C | 1A7Y | 0.73 |  |
DPR | D-PROLINE | A | 1IC9 | 0.73 |  |
DPR | D-PROLINE | A,B | 1JY6 | 0.73 |  |
DPR | D-PROLINE | A | 209D | 0.73 |  |
DPR | D-PROLINE | A | 1BFW | 0.73 |  |
DPR | D-PROLINE | A,B | 1O6I | 0.73 |  |
DPR | D-PROLINE | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P,Q,R, S,T,U,V,W,X | 1UNJ | 0.73 |  |
DPR | D-PROLINE | A,B,C,D | 2R3C | 0.73 |  |
DPR | D-PROLINE | A,B,C,D | 1I3W | 0.73 |  |
DPR | D-PROLINE | A | 316D | 0.73 |  |
DPR | D-PROLINE | A,B,C,D | 1SN9 | 0.73 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5D | 0.73 |  |
DPR | D-PROLINE | A,B | 2Q33 | 0.73 |  |
DPR | D-PROLINE | G,M,P,S | 2I60 | 0.73 |  |
DPR | D-PROLINE | A,B | 1JY4 | 0.73 |  |
DPR | D-PROLINE | P | 1MCF | 0.73 |  |
DPR | D-PROLINE | A,B,C,D,E,F | 1UNM | 0.73 |  |
DPR | D-PROLINE | A | 1ICO | 0.73 |  |
DPR | D-PROLINE | A,B | 1XOF | 0.73 |  |
DPR | D-PROLINE | P | 1MCC | 0.73 |  |
DPR | D-PROLINE | A,B,C,D | 1SNA | 0.73 |  |


