Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00405378
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
BAS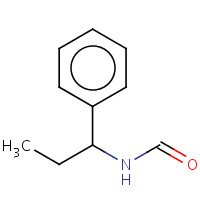 | N-(1-PHENYL-PROPYL)-FORMAMIDE | E,F,G,H | 1NJU | 0.74 |  |
BAS | N-(1-PHENYL-PROPYL)-FORMAMIDE | Y,Z | 1JQ7 | 0.74 |  |
BAS | N-(1-PHENYL-PROPYL)-FORMAMIDE | C | 1NKM | 0.74 |  |
246 | 1-benzyl-3-(2-chloropyridin-4-yl)urea | A | 2QPM | 0.73 |  |
468 | (3S)-N-(3-CHLORO-2-METHYLPHENYL)- 1-CYCLOHEXYL-5-OXOPYRROLIDINE-3- CARBOXAMIDE | A | 2H7P | 0.72 |  |
BAN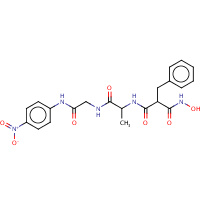 | HONH-BENZYLMALONYL-L-ALANYLGLYCINE- P-NITROANILIDE | A | 5TLN | 0.7 |  |
1MR | N-METHYLANILINE | X | 2OTZ | 0.73 |  |
221 | (2R,3R)-3-{[3,5-BIS(TRIFLUOROMETHYL)PHENYL]AMINO}- 2-CYANO-3-THIOXOPROPANAMIDE | A,B | 2IJN | 0.7 |  |
12Q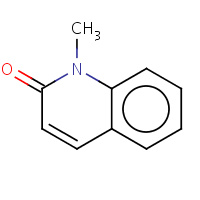 | 1-METHYLQUINOLIN-2(1H)-ONE | A,B | 2F64 | 0.74 |  |
BSU | 1,3-DIPHENYLUREA | A | 3E85 | 0.79 |  |
BSU | 1,3-DIPHENYLUREA | A | 2ZJF | 0.79 |  |
34T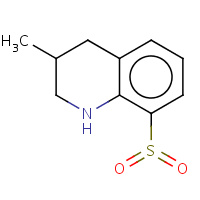 | (3R)-8-(dioxidosulfanyl)-3-methyl- 1,2,3,4-tetrahydroquinoline | H | 1ETR | 0.71 |  |
744 | (3S)-N-(5-CHLORO-2-METHYLPHENYL)- 1-CYCLOHEXYL-5-OXOPYRROLIDINE-3- CARBOXAMIDE | A | 2H7N | 0.72 |  |
264 | (phenylamino)acetonitrile | A | 2RBN | 0.75 |  |
AGB | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA | A | 1EJN | 0.78 |  |
566 | (3S)-1-CYCLOHEXYL-5-OXO-N-PHENYLPYRROLIDINE- 3-CARBOXAMIDE | A | 2H7I | 0.74 |  |
529 | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL- 3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN- 1-YL)PHENYL]PROPANAMIDE | A,C,D | 2BPM | 0.71 |  |
AFF | 2-ACETYLAMINOFLUORENE-3-YL | A | 2GE2 | 0.75 |  |
AU4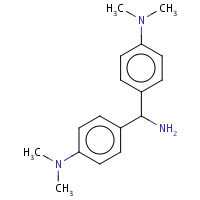 | 4,4'-(AMINOMETHYLENE)BIS(N,N-DIMETHYLANILINE) | A | 2PYZ | 0.8 |  |
BNF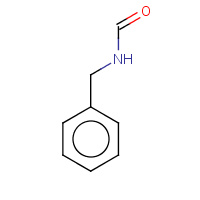 | N-BENZYLFORMAMIDE | A,B | 1U3U | 0.71 |  |
A8B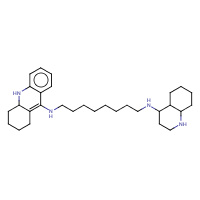 | A | 1ODC | 0.76 |  |


