Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00393174
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
BAS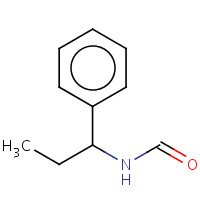 | N-(1-PHENYL-PROPYL)-FORMAMIDE | E,F,G,H | 1NJU | 0.74 |  |
BAS | N-(1-PHENYL-PROPYL)-FORMAMIDE | Y,Z | 1JQ7 | 0.74 |  |
BAS | N-(1-PHENYL-PROPYL)-FORMAMIDE | C | 1NKM | 0.74 |  |
008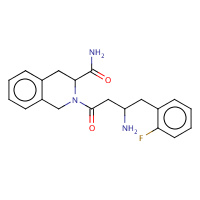 | (S)-2-[(R)-3-AMINO-4-(2-FLUORO- PHENYL)-BUTYRYL]-1,2,3,4-TETRAHYDRO- ISOQUINOLINE-3-CARBOXYLIC ACID AMIDE | A,B,C,D | 2BUC | 0.76 |  |
22U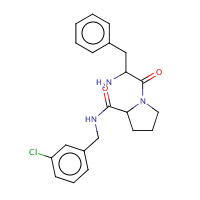 | D-phenylalanyl-N-(3-chlorobenzyl)- L-prolinamide | H,I | 2ZC9 | 0.7 |  |
4FP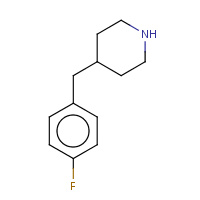 | 4-(4-FLUOROBENZYL)PIPERIDINE | A | 2OHN | 0.71 |  |
37U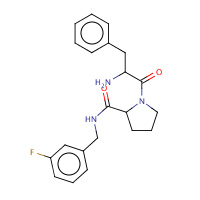 | D-phenylalanyl-N-(3-fluorobenzyl)- L-prolinamide | H,I | 2ZDV | 0.79 |  |
27U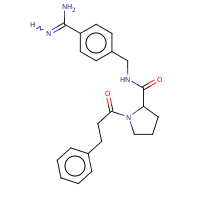 | N-(4-carbamimidoylbenzyl)-1-(3- phenylpropanoyl)-L-prolinamide | H,I | 2ZHQ | 0.71 |  |
BNF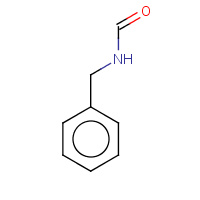 | N-BENZYLFORMAMIDE | A,B | 1U3U | 0.72 |  |
1PC | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE | B,C | 2PCP | 0.72 |  |
F1H | N-(1-benzylpiperidin-4-yl)-4-sulfanylbutanamide | A | 2ZJH | 0.77 |  |
CPU | A,B | 1CR6 | 0.8 |  | |
53U | D-phenylalanyl-N-benzyl-L-prolinamide | H,I | 2ZFF | 0.76 |  |
51U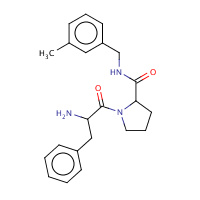 | D-phenylalanyl-N-(3-methylbenzyl)- L-prolinamide | H,I | 2ZF0 | 0.74 |  |
32U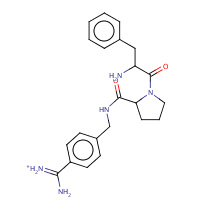 | D-phenylalanyl-N-{4-[amino(iminio)methyl]benzyl}- L-prolinamide | H,I | 2ZDA | 0.71 |  |
998 | N-METHYLALANYL-3-METHYLVALYL-N- (1,2,3,4-TETRAHYDRONAPHTHALEN-1- YL)PROLINAMIDE | A | 1TFQ | 0.71 |  |
12U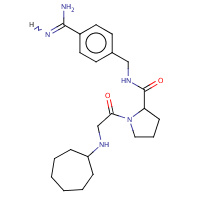 | N-cycloheptylglycyl-N-(4-carbamimidoylbenzyl)- L-prolinamide | A | 2ZHD | 0.71 |  |
12U | N-cycloheptylglycyl-N-(4-carbamimidoylbenzyl)- L-prolinamide | H,I | 2ZHW | 0.71 |  |
12U | N-cycloheptylglycyl-N-(4-carbamimidoylbenzyl)- L-prolinamide | A | 2ZFS | 0.71 |  |
BAC | N-(4-IODO-BENZYL)-FORMAMIDE | A,B,C,D | 2WPO | 0.7 |  |
11U | (S)-N-(4-carbamimidoylbenzyl)-1- (2-(cyclohexylamino)ethanoyl)pyrrolidine- 2-carboxamide | H,I | 3BIV | 0.72 |  |
44U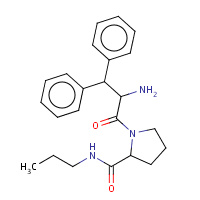 | beta-phenyl-D-phenylalanyl-N-propyl- L-prolinamide | B,D | 3DA9 | 0.7 |  |
F1K | (2S)-4-(4-fluorobenzyl)-N-(3-sulfanylpropyl)piperazine- 2-carboxamide | A,B,C | 2ZJK | 0.76 |  |
F1J | (2S)-4-(4-fluorobenzyl)-N-(2-sulfanylethyl)piperazine- 2-carboxamide | A | 2ZJJ | 0.76 |  |


