Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00022225
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
DUC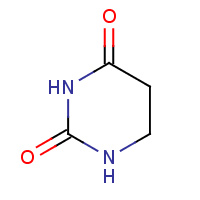 | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE | A,B,C,D | 2FVK | 0.71 |  |
DUC | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE | A,B | 1UAQ | 0.71 |  |
HYN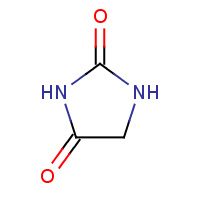 | imidazolidine-2,4-dione | A,B | 3CL7 | 0.85 |  |
QUS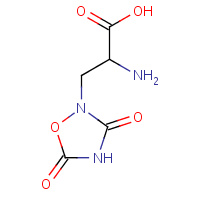 | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A | 1P1O | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A,B | 1S9T | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A,B | 1MM6 | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A | 2JBK | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A,B,C,D,E,F | 2AL4 | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A,B,C | 1MM7 | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A | 3B6T | 0.71 |  |
QUS | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN- 2-YL)-PROPIONIC ACID | A | 2OR4 | 0.71 |  |
175 | 3,5-DIHYDRO-5-METHYLIDENE-4H-IMIDAZOL- 4-ON | A,B,C,D,E,F, G,H | 1T6P | 0.73 |  |
175 | 3,5-DIHYDRO-5-METHYLIDENE-4H-IMIDAZOL- 4-ON | A,B | 1T6J | 0.73 |  |
SNN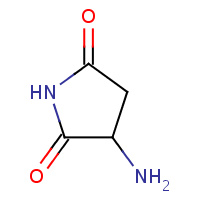 | L-3-AMINOSUCCINIMIDE | A,B | 2IMZ | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A,B | 3C03 | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 3ESM | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A,B | 2OMK | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1AT5 | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1JBE | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1WL8 | 0.7 |  |
3AL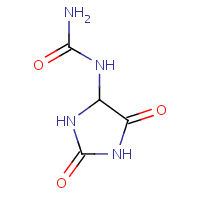 | 1-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4- YL]UREA | A | 2Q37 | 0.85 |  |


