Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00021713
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
AS3 | 4-METHYL-6-PROPYLPYRIDIN-2-AMINE | A,B | 3E6N | 0.76 |  |
LG4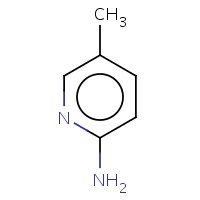 | 5-METHYLPYRIDIN-2-AMINE | A | 2EUP | 0.75 |  |
4AP | 4-AMINOPYRIDINE | A | 1AEG | 0.71 |  |
NTN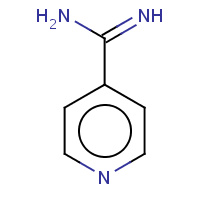 | ISONICOTINAMIDINE | A | 7ADH | 0.71 |  |
JI3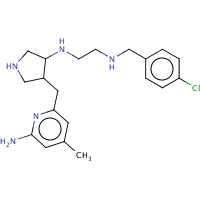 | N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN- 2-YL)METHYL]PYRROLIDIN-3-YL}-N'- (4-CHLOROBENZYL)ETHANE-1,2-DIAMINE | A,B | 3B3O | 0.7 |  |
JI3 | N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN- 2-YL)METHYL]PYRROLIDIN-3-YL}-N'- (4-CHLOROBENZYL)ETHANE-1,2-DIAMINE | A,B | 3DQS | 0.7 |  |
VGD | 6-chloro-1H-benzimidazol-2-amine | A,B,C,D | 2WD7 | 0.72 |  |
CBQ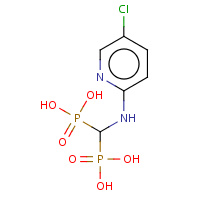 | [(5-CHLORO-PYRIDIN-2-YLAMINO)-PHOSPHONO- METHYL]-PHOSPHONIC ACID | A,B | 1T1S | 0.75 |  |
BVF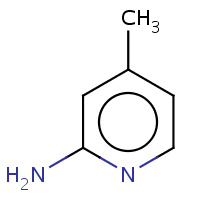 | 4-METHYLPYRIDIN-2-AMINE | A,B | 3E67 | 0.76 |  |
BVF | 4-METHYLPYRIDIN-2-AMINE | A | 2EUT | 0.76 |  |
JI7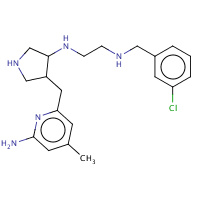 | N-{(3R,4S)-4-[(6-amino-4-methylpyridin- 2-yl)methyl]pyrrolidin-3-yl}-N'- (3-chlorobenzyl)ethane-1,2-diamine | A,B | 3B3P | 0.7 |  |
JI7 | N-{(3R,4S)-4-[(6-amino-4-methylpyridin- 2-yl)methyl]pyrrolidin-3-yl}-N'- (3-chlorobenzyl)ethane-1,2-diamine | A,B | 3DQT | 0.7 |  |
DA1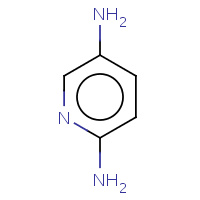 | PYRIDINE-2,5-DIAMINE | A | 2AQD | 0.75 |  |
245 | 1-(2-chloropyridin-4-yl)-3-phenylurea | A | 2QKN | 0.71 |  |
2AP | 2-AMINOPYRIDINE | A | 1AEO | 0.83 |  |
2AQ | QUINOLIN-2-AMINE | A | 2OHL | 0.71 |  |
MPI | IMIDAZO[1,2-A]PYRIDINE | A | 1AEM | 0.7 |  |
1BY | [2-(PYRIDIN-2-YLAMINO)ETHANE-1,1- DIYL]BIS(PHOSPHONIC ACID) | A,B | 2I19 | 0.7 |  |
26D | PYRIDINE-2,6-DIAMINE | A | 2ANZ | 0.87 |  |


