Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03405122
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
EOA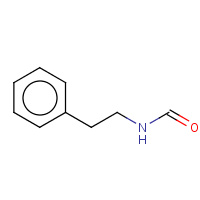 | N-PHENETHYL-FORMAMIDE | H,I | 1A5G | 0.72 |  |
SBD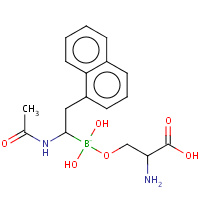 | D-NAPHTHYL-1-ACETAMIDO BORONIC ACID ALANINE | A | 3VSB | 0.81 |  |
NAM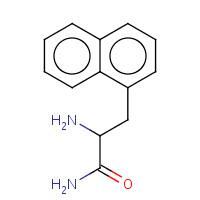 | NAM NAPTHYLAMINOALANINE | A,B,I,J | 3FIV | 0.71 |  |
NAM | NAM NAPTHYLAMINOALANINE | A,B,I,J | 2FIV | 0.71 |  |
CT1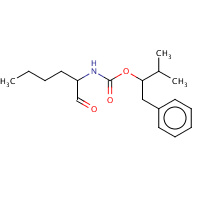 | (1R)-2-METHYL-1-(PHENYLMETHYL)PROPYL[(1S)- 1-FORMYLPENTYL]CARBAMATE | A | 2AUX | 0.7 |  |
SBL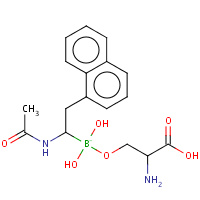 | L-NAPHTHYL-1-ACETAMIDO BORONIC ACID ALANINE | A | 1AV7 | 0.81 |  |
SRD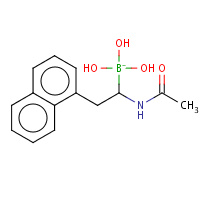 | D-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID | B,C | 4VGC | 0.82 |  |
GCA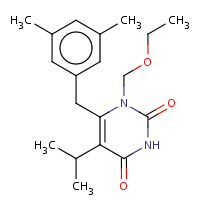 | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL- 5-ISOPROPYLURACIL | A | 1C1B | 0.7 |  |
998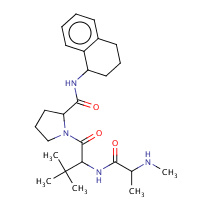 | N-METHYLALANYL-3-METHYLVALYL-N- (1,2,3,4-TETRAHYDRONAPHTHALEN-1- YL)PROLINAMIDE | A | 1TFQ | 0.71 |  |
CPU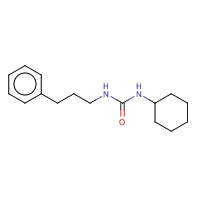 | A,B | 1CR6 | 0.71 |  | |
M1N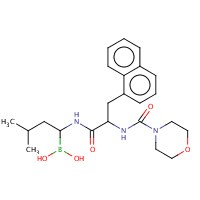 | (1R)-3-METHYL-1-{[N-(MORPHOLIN- 4-YLCARBONYL)-3-(1-NAPHTHYL)-D- ALANYL]AMINO}BUTYLBORONIC ACID | 2,C,E,G,H,J, L,N,P,R,T,V, X,Z | 2FHH | 0.73 |  |
44U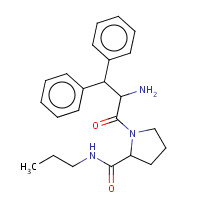 | beta-phenyl-D-phenylalanyl-N-propyl- L-prolinamide | B,D | 3DA9 | 0.7 |  |
SRB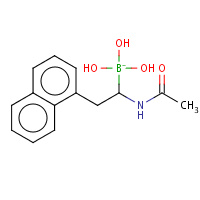 | L-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID | B,C | 3VGC | 0.82 |  |
APE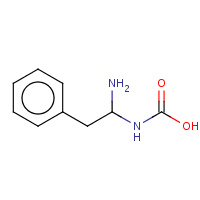 | (1-AMINO-2-PHENYL-ETHYL)-CARBAMIC ACID | E,I | 1SCN | 0.74 |  |


