Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03404111
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
VLM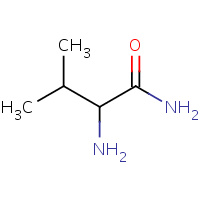 | VALINYLAMINE | G,M,P,S | 1YYM | 0.84 |  |
VLM | VALINYLAMINE | G,M,P,S | 2I5Y | 0.84 |  |
VLM | VALINYLAMINE | G,M,P,S | 1YYL | 0.84 |  |
VLM | VALINYLAMINE | G,M,P,S | 2I60 | 0.84 |  |
HAV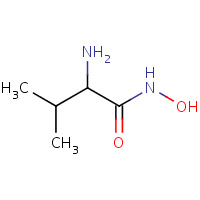 | HYDROXYAMINOVALINE | A | 1BM6 | 0.74 |  |
HAV | HYDROXYAMINOVALINE | A | 1EUB | 0.74 |  |
BUG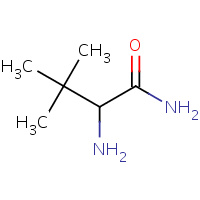 | TERT-LEUCYL AMINE | D | 1D6E | 0.82 |  |
NLN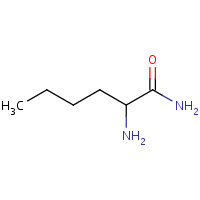 | NORLEUCINE AMIDE | A | 1DW6 | 0.79 |  |
NLN | NORLEUCINE AMIDE | A,B,C | 2AOE | 0.79 |  |
NLN | NORLEUCINE AMIDE | A,B | 1EBK | 0.79 |  |
CLE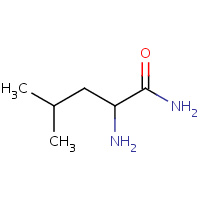 | LEUCINE AMIDE | D | 1D5Z | 0.76 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1QZ0 | 0.76 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1XXV | 0.76 |  |
CLE | LEUCINE AMIDE | A,D | 1D5M | 0.76 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1XXP | 0.76 |  |
CY3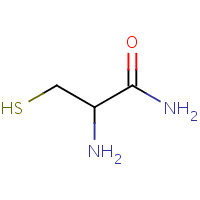 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A | 1D7T | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A | 1DG0 | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | H,I,J,K,L,M | 2V1S | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A,B,C,D | 2V1T | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A | 1DFZ | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | B,F,G,H,I,J | 2BYP | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A | 1R9I | 0.72 |  |
CY3 | 2-AMINO-3-MERCAPTO-PROPIONAMIDE | A | 1DFY | 0.72 |  |
HMA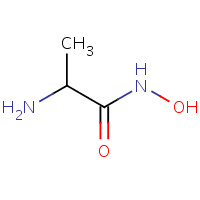 | HYDROXYAMINOALANINE | A | 1AF0 | 0.72 |  |
LYN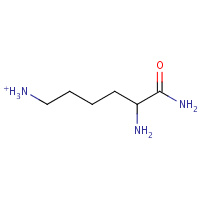 | 2,6-DIAMINO-HEXANOIC ACID AMIDE | A | 1GEA | 0.79 |  |
MNV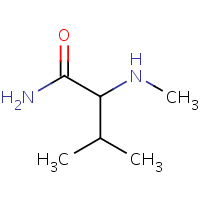 | N-METHYL-C-AMINO VALINE | C | 1CWJ | 0.72 |  |


