Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03211177
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
NIM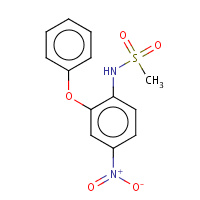 | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE | A | 1ZWP | 0.84 |  |
NIM | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE | A | 3E9X | 0.84 |  |
NIM | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE | A | 2OTH | 0.84 |  |
N4E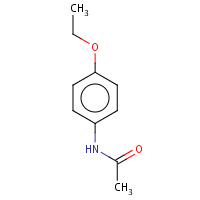 | N-(4-ethoxyphenyl)acetamide | A,B,C,D | 3EBS | 0.76 |  |
MCM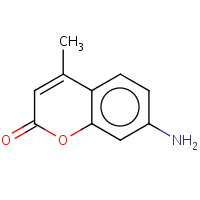 | 7-AMINO-4-METHYL-CHROMEN-2-ONE | A,B,C,D,I,J, K,L | 3EWF | 0.75 |  |
MCM | 7-AMINO-4-METHYL-CHROMEN-2-ONE | A,B,I,L | 2V5W | 0.75 |  |
MCM | 7-AMINO-4-METHYL-CHROMEN-2-ONE | B | 1VDN | 0.75 |  |
MCM | 7-AMINO-4-METHYL-CHROMEN-2-ONE | C | 1VAI | 0.75 |  |
4BG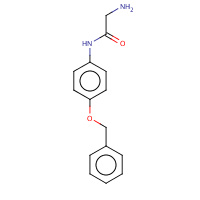 | N-[4-(benzyloxy)phenyl]glycinamide | A | 3CHO | 0.71 |  |
2AF | 2-AMINOPHENOL | A | 1L4N | 0.72 |  |
PNJ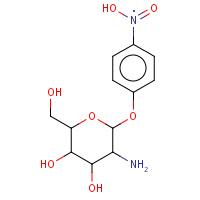 | PNP-BETA-D-GLUCOSAMINE | A,B | 2VZU | 0.71 |  |
PNJ | PNP-BETA-D-GLUCOSAMINE | A,B | 2VZT | 0.71 |  |
RJ6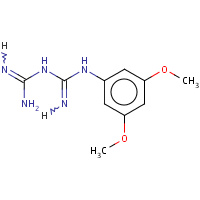 | N-(3,5-dimethoxyphenyl)imidodicarbonimidic diamide | A,B,C,D | 3DG8 | 0.71 |  |
451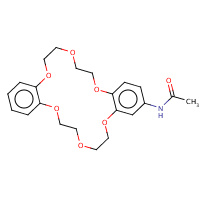 | N-(6,7,9,10,17,18,20,21-octahydrodibenzo[b,k][1,4,7,10,13,16]hexaoxacyclooctadecin- 2-yl)acetamide | A | 3FYX | 0.78 |  |
4BS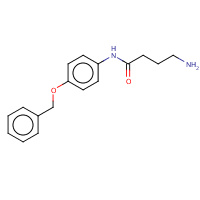 | 4-amino-N-[4-(benzyloxy)phenyl]butanamide | A | 3CHR | 0.71 |  |
EPN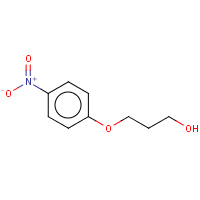 | 3-(4-NITRO-PHENOXY)-PROPAN-1-OL | A | 2SAM | 0.72 |  |
GAT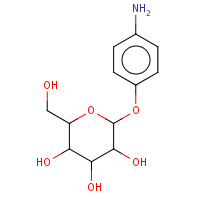 | 4'-AMINOPHENYL-ALPHA-D-GALACTOPYRANOSIDE | D,E,F,G,H | 1EFI | 0.72 |  |


