Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03208320
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
AIO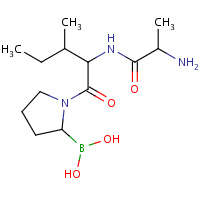 | [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN- 2-YL]BORONIC ACID | A | 2EEP | 0.75 |  |
AIO | [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN- 2-YL]BORONIC ACID | A | 2Z3Z | 0.75 |  |
PPL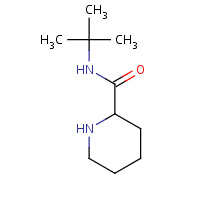 | PIPERIDINE-2-CARBOXYLIC ACID TERT- BUTYLAMIDE | A,B | 1IDB | 0.7 |  |
PPL | PIPERIDINE-2-CARBOXYLIC ACID TERT- BUTYLAMIDE | A,B | 1IDA | 0.7 |  |
LPD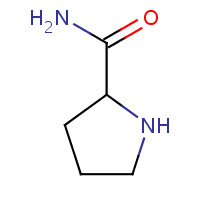 | L-PROLINAMIDE | H,S | 2H9E | 0.76 |  |
GIO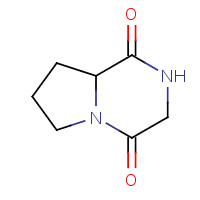 | CYCLO-(GLYCINE-L-PROLINE) INHIBITOR | A,B | 1W1P | 0.84 |  |
BPR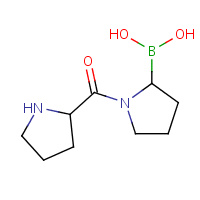 | (2R)-N-[(2R)-2-(DIHYDROXYBORYL)- 1-L-PROLYLPYRROLIDIN-2-YL]-N-[(5R)- 5-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN- 2-YL]-L-PROLINAMIDE | A,B,C,D | 2AJD | 0.78 |  |
CGN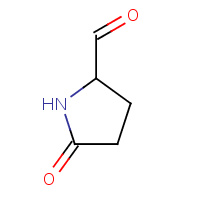 | 5-OXO-PYRROLIDINE-2-CARBALDEHYDE | A | 3CAR | 0.73 |  |
CGN | 5-OXO-PYRROLIDINE-2-CARBALDEHYDE | A | 1I3U | 0.73 |  |
CGN | 5-OXO-PYRROLIDINE-2-CARBALDEHYDE | A | 3CAO | 0.73 |  |
SNN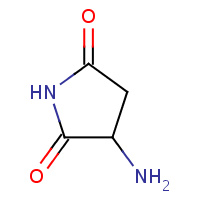 | L-3-AMINOSUCCINIMIDE | A,B | 2IMZ | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A,B | 3C03 | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 3ESM | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A,B | 2OMK | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1AT5 | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1JBE | 0.7 |  |
SNN | L-3-AMINOSUCCINIMIDE | A | 1WL8 | 0.7 |  |
ALJ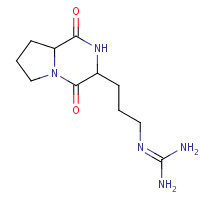 | CYCLO-(L-ARGININE-L-PROLINE) INHIBITOR | A,B | 1W1V | 0.76 |  |


