Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03175965
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
HQ6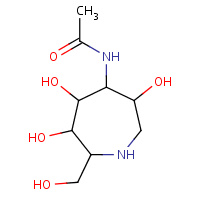 | N-[(3R,4S,5R,6R,7R)-3,5,6-trihydroxy- 7-(hydroxymethyl)azepan-4-yl]acetamide | A,B | 2W66 | 0.7 |  |
NAU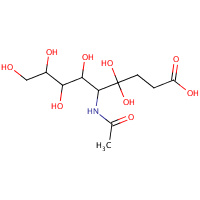 | 4,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDONONANOIC ACID | A,C | 1F7B | 0.74 |  |
MG8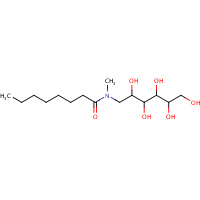 | N-OCTANOYL-N-METHYLGLUCAMINE | A,B | 1SMH | 0.7 |  |
MG8 | N-OCTANOYL-N-METHYLGLUCAMINE | A | 1Q61 | 0.7 |  |
MG8 | N-OCTANOYL-N-METHYLGLUCAMINE | A | 1Q8U | 0.7 |  |
MG8 | N-OCTANOYL-N-METHYLGLUCAMINE | A | 1SVE | 0.7 |  |
DL6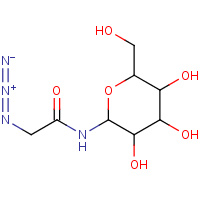 | 2-AZIDO-N-((2R,3R,4S,5S,6R)-3,4,5- TRIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO- 2H-PYRAN-2-YL)ACETAMIDE | A | 2FFR | 0.72 |  |
CR1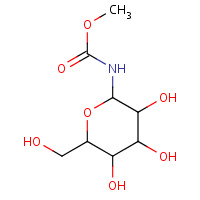 | 1-DEOXY-1-METHOXYCARBAMIDO-BETA- D-GLUCOPYRANOSE | A | 1FU7 | 0.75 |  |
H4P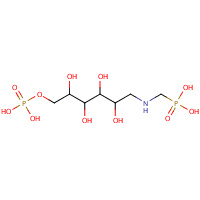 | 1-DEOXY-6-O-PHOSPHONO-1-[(PHOSPHONOMETHYL)AMINO]- L-THREO-HEXITOL | A,B | 1PCW | 0.75 |  |
233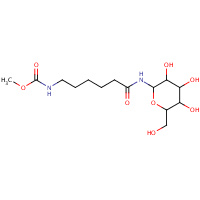 | [5-(3,4,5-TRIHYDROXY-6-HYDROXYMETHYL- TETRAHYDRO-PYRAN-2-YLCARBAMOYL)- PENTYL]-CARBAMIC ACID METHYL ESTER | D,E,F,G,H | 1MD2 | 0.74 |  |
8GP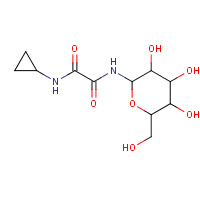 | A | 2F3U | 0.71 |  | |
DQQ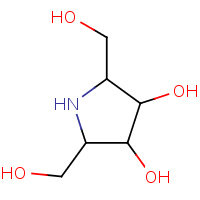 | 2,5-DIDEOXY-2,5-IMINO-D-MANNITOL | A | 2AEY | 0.7 |  |
NBG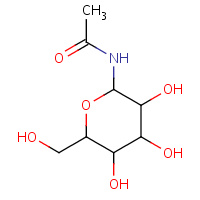 | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1L5Q | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3CEM | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1L5R | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1EXV | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1L5S | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1L7X | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A | 1WW2 | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A | 2PRJ | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1FC0 | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3CEJ | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3CEH | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1XOI | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3DDW | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 1EM6 | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3DDS | 0.74 |  |
NBG | 1-N-ACETYL-BETA-D-GLUCOSAMINE | A,B | 3DD1 | 0.74 |  |
4GP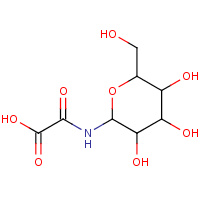 | N-(BETA-D-GLUCOPYRANOSYL)OXAMIC ACID | A | 2F3P | 0.81 |  |
DMJ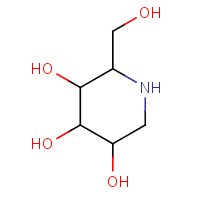 | 1-DEOXYMANNOJIRIMYCIN | A,B | 1KRE | 0.74 |  |
DMJ | 1-DEOXYMANNOJIRIMYCIN | A | 1FO2 | 0.74 |  |
DMJ | 1-DEOXYMANNOJIRIMYCIN | A | 1G6I | 0.74 |  |
DMJ | 1-DEOXYMANNOJIRIMYCIN | A | 1HXK | 0.74 |  |
7GP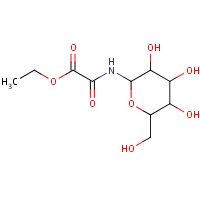 | ETHYL-N-(BETA-D-GLUCOPYRANOSYL)OXAMATE | A | 2F3S | 0.79 |  |
HMN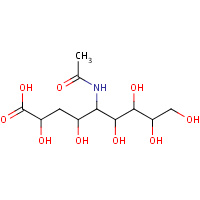 | 2,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDO NONANOIC ACID | A,B,C,D | 1F73 | 0.72 |  |
DIG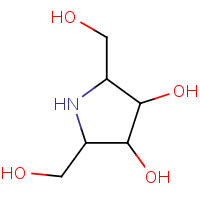 | 2,5-DIDEOXY-2,5-IMINO-D-GLUCITOL | A,B | 1DID | 0.7 |  |
AMU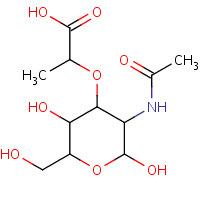 | BETA-N-ACETYLMURAMIC ACID | A,B,C,D | 2APH | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | A,P | 1TWQ | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | A | 1D0K | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | S | 9LYZ | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | E,S | 148L | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | D,K | 3CYQ | 0.71 |  |
AMU | BETA-N-ACETYLMURAMIC ACID | U | 2AIZ | 0.71 |  |
6GP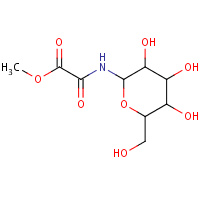 | METHYL-N-(BETA-D-GLUCOPYRANOSYL)OXAMATE | A | 2F3Q | 0.78 |  |
NA1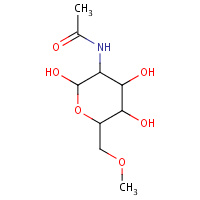 | METHYL N-ACETYL ALLOSAMINE | A | 1HKJ | 0.71 |  |
NA1 | METHYL N-ACETYL ALLOSAMINE | A,B | 3FY1 | 0.71 |  |
NA1 | METHYL N-ACETYL ALLOSAMINE | A | 1HKI | 0.71 |  |
MMN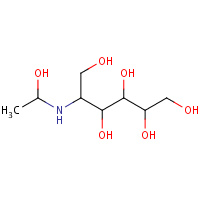 | 5-DEOXY-5-{[(1S)-1-HYDROXYETHYL]AMINO}- D-GLUCITOL | A | 1XUZ | 0.8 |  |
MUB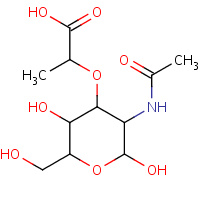 | N-ACETYLMURAMIC ACID | L,N | 1WCO | 0.71 |  |


