Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03090154
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
TN7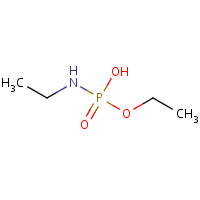 | ethyl hydrogen ethylamidophosphate | A | 2WIJ | 0.73 |  |
OPE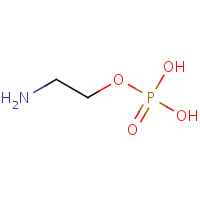 | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A,B,C | 2IQX | 0.85 |  |
OPE | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P,Q,R | 2HIL | 0.85 |  |
OPE | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A | 2HI2 | 0.85 |  |
OPE | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A | 3FI8 | 0.85 |  |
OPE | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A,B | 3FLT | 0.85 |  |
OPE | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | A,B | 1B7A | 0.85 |  |
GPE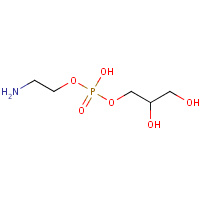 | L-ALPHA-GLYCEROPHOSPHORYLETHANOLAMINE | A | 1A8B | 0.74 |  |
PSE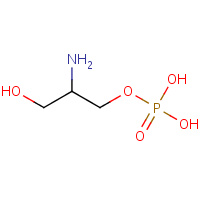 | O-PHOSPHOETHANOLAMINE | A | 2R7B | 0.79 |  |
PSE | O-PHOSPHOETHANOLAMINE | A,B | 1A25 | 0.79 |  |
PSE | O-PHOSPHOETHANOLAMINE | P,Q | 1A37 | 0.79 |  |
PSE | O-PHOSPHOETHANOLAMINE | A,B | 2FAE | 0.79 |  |
EA2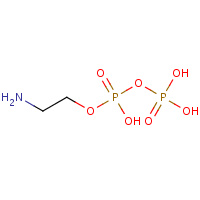 | AMINOETHANOLPYROPHOSPHATE | A | 2FCP | 0.83 |  |
EA2 | AMINOETHANOLPYROPHOSPHATE | A,B | 2I6K | 0.83 |  |
EA2 | AMINOETHANOLPYROPHOSPHATE | A | 1FCP | 0.83 |  |
TC5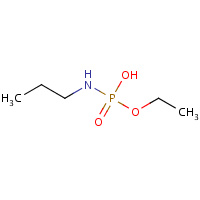 | ETHYL HYDROGEN PROPYLAMIDOPHOSPHATE | A | 2WIK | 0.71 |  |
210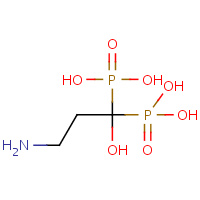 | PAMIDRONATE | F | 2F89 | 0.76 |  |
GG7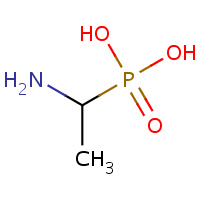 | [(1R)-1-AMINOETHYL]PHOSPHONIC ACID | A,B,C,D | 2PBK | 0.74 |  |
AHD | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE | A,B,C | 1YHM | 0.7 |  |
AHD | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE | F | 2F92 | 0.7 |  |


