Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02856823
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
DXP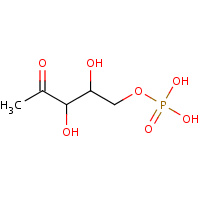 | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE | B,C,D,E,F,G,H | 3GK0 | 0.75 |  |
DXP | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE | A,B,C,E,F,G | 1M5W | 0.75 |  |
DXP | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE | A,B,C,D | 1IXN | 0.75 |  |
DXP | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE | A,B | 1Q0Q | 0.75 |  |
3MF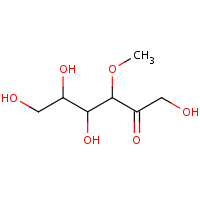 | 3-O-METHYLFRUCTOSE IN LINEAR FORM | A,B | 1XYC | 0.75 |  |
KDG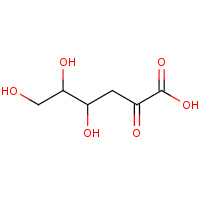 | 2-KETO-3-DEOXYGLUCONATE | A,B | 1V1A | 0.71 |  |
KDG | 2-KETO-3-DEOXYGLUCONATE | A,B,C | 2VAR | 0.71 |  |
KDG | 2-KETO-3-DEOXYGLUCONATE | A,B,C,D | 2QJN | 0.71 |  |
FUD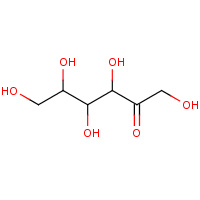 | D-fructose | A,B,C,D | 2QUN | 0.78 |  |
TAG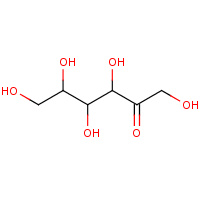 | D-tagatose | A,B,C,D | 2QUM | 0.78 |  |
GLO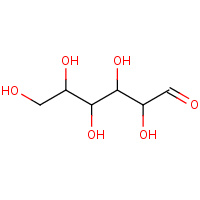 | D-GLUCOSE IN LINEAR FORM | A,B | 1XYB | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FQD | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A,B | 1XYM | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A,B | 1EZ9 | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FBO | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1AC0 | 0.7 |  |
GLO | D-GLUCOSE IN LINEAR FORM | A | 1FQC | 0.7 |  |
DMV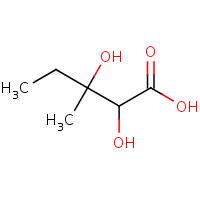 | 2,3-DIHYDROXY-VALERIANIC ACID | A,B,C,D | 1QMG | 0.71 |  |
LFC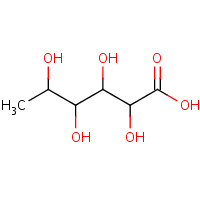 | 6-DEOXY-L-GALACTONIC ACID | A | 2HXU | 0.71 |  |
AOS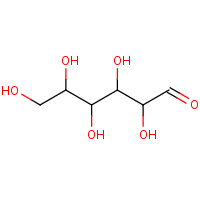 | D-ALLOSE | A,B,C,D | 2I57 | 0.7 |  |
I1N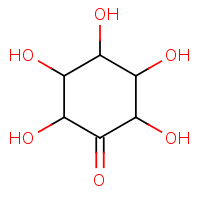 | (2S,3R,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXANONE | A,B | 2IBN | 0.72 |  |
XLS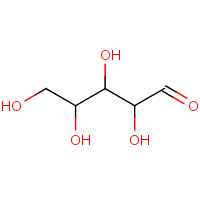 | D-XYLOSE (LINEAR FORM) | A,B | 2QW5 | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 2BRP | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 3XIS | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 8XIM | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 4XIS | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 9XIM | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 5XIN | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 8XIA | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A,B,C,D | 6XIM | 0.76 |  |
XLS | D-XYLOSE (LINEAR FORM) | A | 1XIC | 0.76 |  |
DNO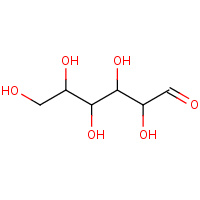 | D-mannose | A | 3BDK | 0.7 |  |
RNS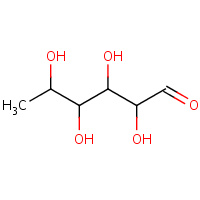 | L-RHAMNOSE | A,B,C,D | 1X8D | 0.76 |  |
RNS | L-RHAMNOSE | A,B,C,D | 1DE6 | 0.76 |  |
RNS | L-RHAMNOSE | A,B,C,D | 2I56 | 0.76 |  |
XUL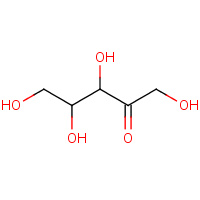 | D-XYLULOSE | A | 3CWH | 0.79 |  |
XUL | D-XYLULOSE | A | 1XII | 0.79 |  |
XUL | D-XYLULOSE | A,B | 2ITM | 0.79 |  |


