Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02389874
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
GCN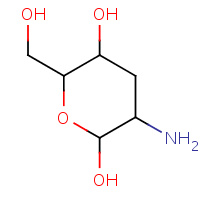 | 3-DEOXY-D-GLUCOSAMINE | A | 1FI1 | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFG | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QKC | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A,B | 2GRX | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QJQ | 0.71 |  |
GCN | 3-DEOXY-D-GLUCOSAMINE | A | 1QFF | 0.71 |  |
GRF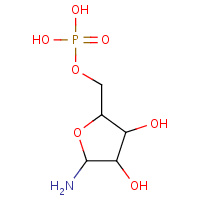 | 5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE | A | 1ZLY | 0.75 |  |
1GN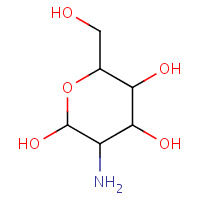 | 2-DEOXY-2-AMINOGALACTOSE | A,B | 3GAL | 0.71 |  |
GOO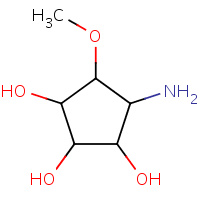 | (1R,2R,3R,4S,5R)-4-amino-5-methoxycyclopentane- 1,2,3-triol | A | 3DX4 | 0.74 |  |
AGL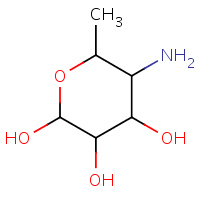 | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1CPU | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1NM9 | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1MFV | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1PIG | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | X | 1Z32 | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 1MFU | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 3BLK | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | X | 3BLP | 0.7 |  |
AGL | 4,6-DIDEOXY-4-AMINO-ALPHA-D-GLUCOSE | A | 3DHP | 0.7 |  |
PA1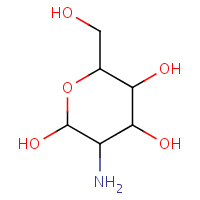 | PAROMOMYCIN (RING 1) | A | 1PBR | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QKC | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFG | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1FI1 | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A,B,C,D | 3FXI | 0.71 |  |
PA1 | PAROMOMYCIN (RING 1) | A | 1QFF | 0.71 |  |
GCS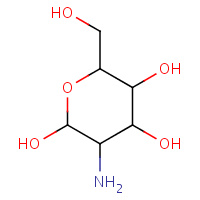 | D-GLUCOSAMINE | A | 3CO4 | 0.71 |  |
GCS | D-GLUCOSAMINE | A | 1QGI | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZS | 0.71 |  |
GCS | D-GLUCOSAMINE | A | 1E9L | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B,C,D | 3FXI | 0.71 |  |
GCS | D-GLUCOSAMINE | A,B | 2VZV | 0.71 |  |
DAG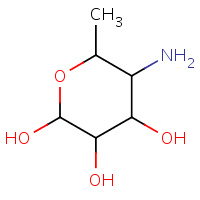 | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A | 2PIK | 0.7 |  |
DAG | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A | 6CGT | 0.7 |  |
DAG | 4,6-DIDEOXY-4-AMINO-BETA-D-GLUCOPYRANOSIDE | A,B | 1PIK | 0.7 |  |


