Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02355444
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
MVA | N-METHYLVALINE | A | 209D | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C,D,E,F, G,H | 3BO7 | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWH | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P,Q,R, S,T,U,V,W,X | 1UNJ | 0.72 |  |
MVA | N-METHYLVALINE | A | 1DSD | 0.72 |  |
MVA | N-METHYLVALINE | B | 1L1V | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C,D,E,F, I,J,K,L,M,N | 2ESL | 0.72 |  |
MVA | N-METHYLVALINE | B,D,F,H,J,L, N,P,R,T | 2RMB | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWO | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWL | 0.72 |  |
MVA | N-METHYLVALINE | B | 1MIK | 0.72 |  |
MVA | N-METHYLVALINE | A | 1CSA | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWM | 0.72 |  |
MVA | N-METHYLVALINE | A | 1PFE | 0.72 |  |
MVA | N-METHYLVALINE | A,B,D,E,F,H | 1M63 | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWC | 0.72 |  |
MVA | N-METHYLVALINE | B,D,F,H | 2RMC | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWK | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CYN | 0.72 |  |
MVA | N-METHYLVALINE | C,D | 1MNV | 0.72 |  |
MVA | N-METHYLVALINE | C,D | 2OJU | 0.72 |  |
MVA | N-METHYLVALINE | B,D,F,H,J,L, N,P,R,T | 2RMA | 0.72 |  |
MVA | N-METHYLVALINE | A | 1CYB | 0.72 |  |
MVA | N-METHYLVALINE | D | 1QNG | 0.72 |  |
MVA | N-METHYLVALINE | A,C,E,G | 2ADW | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWB | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C,D,E,F | 1UNM | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 1OVF | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 185D | 0.72 |  |
MVA | N-METHYLVALINE | A | 1XVK | 0.72 |  |
MVA | N-METHYLVALINE | A,C | 1XVR | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 173D | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 1FJA | 0.72 |  |
MVA | N-METHYLVALINE | B | 3CYS | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C | 1A7Y | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C | 1L5G | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C,D | 1I3W | 0.72 |  |
MVA | N-METHYLVALINE | A | 316D | 0.72 |  |
MVA | N-METHYLVALINE | A | 1XVN | 0.72 |  |
MVA | N-METHYLVALINE | C | 1IKF | 0.72 |  |
MVA | N-METHYLVALINE | A,M,N | 2Z6W | 0.72 |  |
MVA | N-METHYLVALINE | A,B,C | 1QFI | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 3GO3 | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWA | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWF | 0.72 |  |
MVA | N-METHYLVALINE | B,D,F,H,J,L, N,P | 1C5F | 0.72 |  |
MVA | N-METHYLVALINE | D,E,F | 1XQ7 | 0.72 |  |
MVA | N-METHYLVALINE | A | 1CYA | 0.72 |  |
MVA | N-METHYLVALINE | A,B,D | 1MF8 | 0.72 |  |
MVA | N-METHYLVALINE | A,B | 1A7Z | 0.72 |  |
MVA | N-METHYLVALINE | A | 1DSC | 0.72 |  |
MVA | N-METHYLVALINE | T,U,V | 2POY | 0.72 |  |
MVA | N-METHYLVALINE | A | 2D55 | 0.72 |  |
MVA | N-METHYLVALINE | C | 1BCK | 0.72 |  |
MVA | N-METHYLVALINE | C | 1CWI | 0.72 |  |
MVA | N-METHYLVALINE | C,D | 1QNH | 0.72 |  |
MVA | N-METHYLVALINE | A | 1VS2 | 0.72 |  |
LLY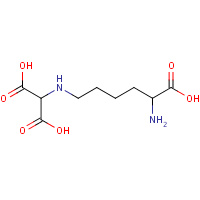 | NZ-(DICARBOXYMETHYL)LYSINE | A,B | 1UCW | 0.71 |  |
PAU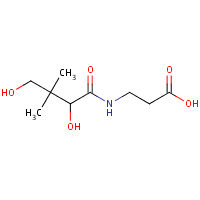 | PANTOTHENOIC ACID | A,B | 2F9W | 0.71 |  |
PAU | PANTOTHENOIC ACID | A,B,C,D,E,F | 3BF1 | 0.71 |  |
PAU | PANTOTHENOIC ACID | A,B,C,D | 1SQ5 | 0.71 |  |
PAU | PANTOTHENOIC ACID | A,B,C,D,E,F | 3BEX | 0.71 |  |


