Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02325946
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
MCL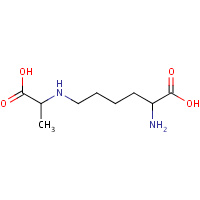 | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 1O5K | 0.79 |  |
MCL | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 3CPR | 0.79 |  |
MCL | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 2VC6 | 0.79 |  |
SHR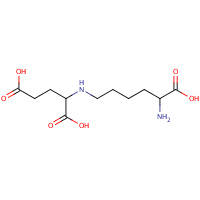 | N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID | A,B,C,D,E,F, G,H | 1E5Q | 0.73 |  |
140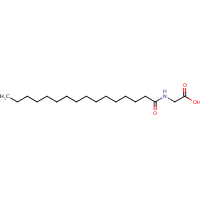 | N-PALMITOYLGLYCINE | A,B | 1JPZ | 0.71 |  |
140 | N-PALMITOYLGLYCINE | A,B | 1ZOA | 0.71 |  |
140 | N-PALMITOYLGLYCINE | A,B | 3CBD | 0.71 |  |
SUO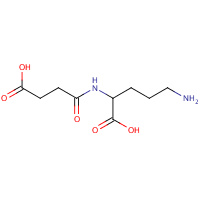 | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE | A,B,C,D | 1YNH | 0.71 |  |
LDH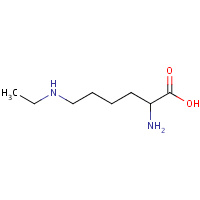 | N~6~-ETHYL-L-LYSINE | A,K | 2IOF | 0.73 |  |
MHA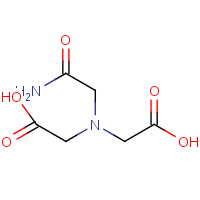 | (CARBAMOYLMETHYL-CARBOXYMETHYL- AMINO)-ACETIC ACID | A,B | 2IXX | 0.71 |  |
MHA | (CARBAMOYLMETHYL-CARBOXYMETHYL- AMINO)-ACETIC ACID | A,B | 1IX1 | 0.71 |  |
NMC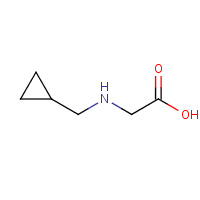 | N-CYCLOPROPYLMETHYL GLYCINE | C,D | 3SEM | 0.79 |  |
DLS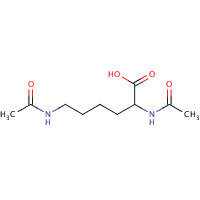 | DI-ACETYL-LYSINE | A,B,C,D,E,F | 1FVM | 0.73 |  |
LLY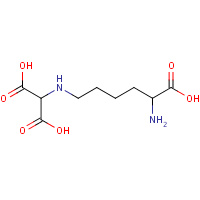 | NZ-(DICARBOXYMETHYL)LYSINE | A,B | 1UCW | 0.75 |  |
DPR | D-PROLINE | P | 1MCB | 0.7 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5B | 0.7 |  |
DPR | D-PROLINE | A | 2KDQ | 0.7 |  |
DPR | D-PROLINE | P | 1MCS | 0.7 |  |
DPR | D-PROLINE | A,B | 2AXI | 0.7 |  |
DPR | D-PROLINE | A | 1JY9 | 0.7 |  |
DPR | D-PROLINE | A | 2JUE | 0.7 |  |
DPR | D-PROLINE | P | 1MCK | 0.7 |  |
DPR | D-PROLINE | P | 1MCR | 0.7 |  |
DPR | D-PROLINE | A | 2D55 | 0.7 |  |
DPR | D-PROLINE | A | 1T8J | 0.7 |  |
DPR | D-PROLINE | A,B | 2EJ6 | 0.7 |  |
DPR | D-PROLINE | P | 1MCE | 0.7 |  |
DPR | D-PROLINE | P | 1MCI | 0.7 |  |
DPR | D-PROLINE | A,B | 1SNE | 0.7 |  |
DPR | D-PROLINE | A,B,C | 1QFI | 0.7 |  |
DPR | D-PROLINE | A,P | 1MCD | 0.7 |  |
DPR | D-PROLINE | A | 1ICL | 0.7 |  |
DPR | D-PROLINE | P | 1MCQ | 0.7 |  |
DPR | D-PROLINE | G,M,P,S | 2I5Y | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 3BOG | 0.7 |  |
DPR | D-PROLINE | A,L | 1ZEA | 0.7 |  |
DPR | D-PROLINE | A,B | 173D | 0.7 |  |
DPR | D-PROLINE | A,P | 1MCJ | 0.7 |  |
DPR | D-PROLINE | A,B,C | 1A7Y | 0.7 |  |
DPR | D-PROLINE | A | 1IC9 | 0.7 |  |
DPR | D-PROLINE | A,B | 1JY6 | 0.7 |  |
DPR | D-PROLINE | A | 209D | 0.7 |  |
DPR | D-PROLINE | A | 1BFW | 0.7 |  |
DPR | D-PROLINE | A,B | 1O6I | 0.7 |  |
DPR | D-PROLINE | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P,Q,R, S,T,U,V,W,X | 1UNJ | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 2R3C | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1I3W | 0.7 |  |
DPR | D-PROLINE | A | 316D | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1SN9 | 0.7 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5D | 0.7 |  |
DPR | D-PROLINE | A,B | 2Q33 | 0.7 |  |
DPR | D-PROLINE | G,M,P,S | 2I60 | 0.7 |  |
DPR | D-PROLINE | A,B | 1JY4 | 0.7 |  |
DPR | D-PROLINE | P | 1MCF | 0.7 |  |
DPR | D-PROLINE | A,B,C,D,E,F | 1UNM | 0.7 |  |
DPR | D-PROLINE | A | 1ICO | 0.7 |  |
DPR | D-PROLINE | A,B | 1XOF | 0.7 |  |
DPR | D-PROLINE | P | 1MCC | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1SNA | 0.7 |  |
NC3 | N-[(CYCLOHEXYLAMINO)CARBONYL]GLYCINE | P | 1ZD2 | 0.75 |  |
EDT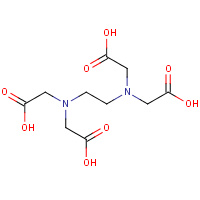 | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A | 2AXN | 0.71 |  |
EDT | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A | 1NNF | 0.71 |  |
EDT | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A,B | 1ZLQ | 0.71 |  |
IPG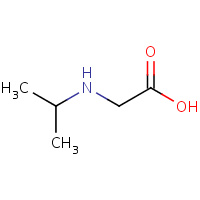 | N-ISOPROPYL GLYCINE | C,D | 2SEM | 0.77 |  |
CL5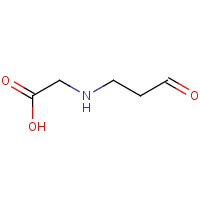 | N-(3-OXOPROPYL)GLYCINE | A | 2IOT | 0.74 |  |
LYP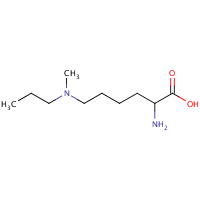 | N~6~-METHYL-N~6~-PROPYL-L-LYSINE | A,E | 2UXN | 0.7 |  |
MAA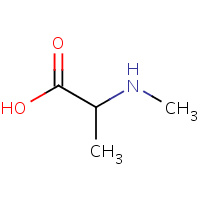 | N-METHYLALANINE | A,B | 1A7Z | 0.71 |  |
MAA | N-METHYLALANINE | A,B | 2VSL | 0.71 |  |
MAA | N-METHYLALANINE | C | 1CWI | 0.71 |  |
MAA | N-METHYLALANINE | C,F,G,H | 3DW8 | 0.71 |  |
MAA | N-METHYLALANINE | A,B,C,D,E,F, G,H | 2NYL | 0.71 |  |
MAA | N-METHYLALANINE | D | 1D5X | 0.71 |  |
MAA | N-METHYLALANINE | C | 1PYW | 0.71 |  |
MAA | N-METHYLALANINE | A,B,C,D,E,F, G,H | 2NYM | 0.71 |  |
MAA | N-METHYLALANINE | C,I | 2IE3 | 0.71 |  |
MAA | N-METHYLALANINE | C,F,X,Y | 2NPP | 0.71 |  |


