Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02234985
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
EDT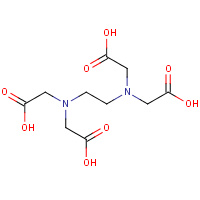 | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A | 2AXN | 0.72 |  |
EDT | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A | 1NNF | 0.72 |  |
EDT | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]- CARBOXYMETHYL-AMINO}-ACETIC ACID | A,B | 1ZLQ | 0.72 |  |
HIO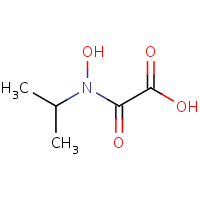 | N-HYDROXY-N-ISOPROPYLOXAMIC ACID | I,J,K,L | 1YVE | 0.7 |  |
HDA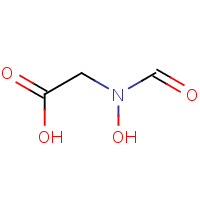 | HADACIDIN | A | 1CIB | 0.85 |  |
HDA | HADACIDIN | A | 1CG1 | 0.85 |  |
HDA | HADACIDIN | A | 1CG0 | 0.85 |  |
HDA | HADACIDIN | A | 1P9B | 0.85 |  |
HDA | HADACIDIN | A | 1CH8 | 0.85 |  |
HDA | HADACIDIN | A | 1JUY | 0.85 |  |
HDA | HADACIDIN | A | 1CG3 | 0.85 |  |
HDA | HADACIDIN | A | 1LON | 0.85 |  |
CMS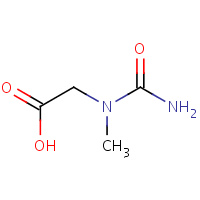 | CARBAMOYL SARCOSINE | A,B | 1CHM | 0.73 |  |
HAD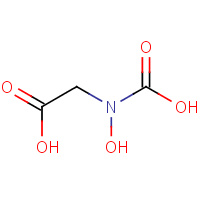 | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1KKB | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1GIM | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1GIN | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1KSZ | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1NHT | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 1KKF | 0.79 |  |
HAD | (CARBOXYHYDROXYAMINO)ETHANOIC ACID | A | 2GCQ | 0.79 |  |
AAC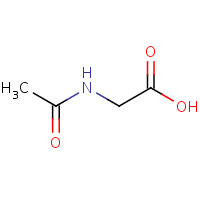 | ACETYLAMINO-ACETIC ACID | A,B | 1NG3 | 0.74 |  |
AAC | ACETYLAMINO-ACETIC ACID | A | 1QD8 | 0.74 |  |
MHA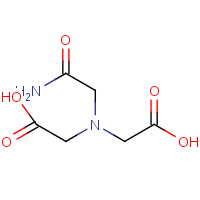 | (CARBAMOYLMETHYL-CARBOXYMETHYL- AMINO)-ACETIC ACID | A,B | 2IXX | 0.73 |  |
MHA | (CARBAMOYLMETHYL-CARBOXYMETHYL- AMINO)-ACETIC ACID | A,B | 1IX1 | 0.73 |  |
BET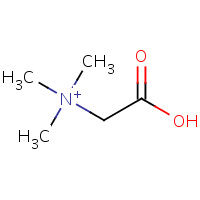 | TRIMETHYL GLYCINE | A | 1RCC | 0.76 |  |
BET | TRIMETHYL GLYCINE | A,B | 3DSB | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1RCI | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1SW2 | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1RCE | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1R9L | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1RCD | 0.76 |  |
BET | TRIMETHYL GLYCINE | A,B,C | 2WIT | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 2B4L | 0.76 |  |
BET | TRIMETHYL GLYCINE | A | 1RCG | 0.76 |  |
BET | TRIMETHYL GLYCINE | A,B,C,D | 1WWJ | 0.76 |  |
DMG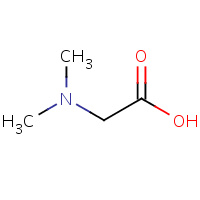 | N,N-DIMETHYLGLYCINE | A,B,C,D | 1VRQ | 0.76 |  |
DMG | N,N-DIMETHYLGLYCINE | A,B,C,D | 1X31 | 0.76 |  |
DMG | N,N-DIMETHYLGLYCINE | A,B,C | 1XKP | 0.76 |  |
DMG | N,N-DIMETHYLGLYCINE | A,B | 1EL5 | 0.76 |  |
NTA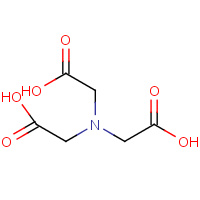 | NITRILOTRIACETIC ACID | A | 1GVC | 0.84 |  |
NTA | NITRILOTRIACETIC ACID | A | 1NFT | 0.84 |  |
BCN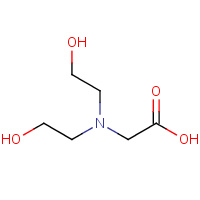 | BICINE | A,B,C,D | 2V8H | 0.82 |  |
BCN | BICINE | A | 1QUS | 0.82 |  |
BCN | BICINE | A | 1LTM | 0.82 |  |
BCN | BICINE | A | 2JC5 | 0.82 |  |
BCN | BICINE | A,B,C,D | 1V0J | 0.82 |  |
BCN | BICINE | A | 1QDR | 0.82 |  |
BCN | BICINE | A | 1KI0 | 0.82 |  |
BCN | BICINE | Y,Z | 1KMI | 0.82 |  |
BCN | BICINE | A,B,C | 2OV5 | 0.82 |  |
BCN | BICINE | A | 2A81 | 0.82 |  |
BCN | BICINE | A,B,C,D | 2V8G | 0.82 |  |
BCN | BICINE | A,B | 3HWR | 0.82 |  |
BCN | BICINE | A | 2R6S | 0.82 |  |
BCN | BICINE | A,B | 2R4J | 0.82 |  |
OGA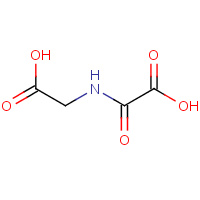 | N-OXALYOLGLYCINE | A,B,C,D | 2OQ6 | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A | 2RDS | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A | 1H2M | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A | 2QRL | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,C,D | 2OX0 | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A | 2RDR | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,C,D | 2OS2 | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,I,J | 2PXJ | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,F,G | 2Q8E | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,I,J | 2P5B | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A | 1H2K | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B,C,D | 2OT7 | 0.77 |  |
OGA | N-OXALYOLGLYCINE | A,B | 2OQ7 | 0.77 |  |


