Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs00482813
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
DNL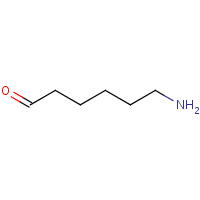 | 6-AMINO-HEXANAL | E,F,G,H | 1NJU | 0.73 |  |
DNL | 6-AMINO-HEXANAL | C | 1NKM | 0.73 |  |
CLE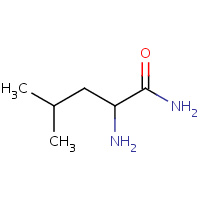 | LEUCINE AMIDE | D | 1D5Z | 0.83 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1QZ0 | 0.83 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1XXV | 0.83 |  |
CLE | LEUCINE AMIDE | A,D | 1D5M | 0.83 |  |
CLE | LEUCINE AMIDE | C,D,E,F | 1XXP | 0.83 |  |
BUG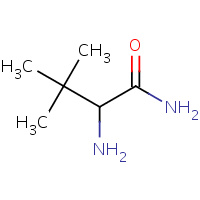 | TERT-LEUCYL AMINE | D | 1D6E | 0.76 |  |
LYM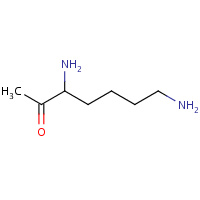 | DEOXY-METHYL-LYSINE | I | 4PAD | 0.8 |  |
LYM | DEOXY-METHYL-LYSINE | A,C,D | 1S4V | 0.8 |  |
LYM | DEOXY-METHYL-LYSINE | A,B,C,D | 2ID4 | 0.8 |  |
VLM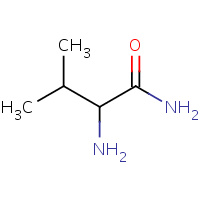 | VALINYLAMINE | G,M,P,S | 1YYM | 0.78 |  |
VLM | VALINYLAMINE | G,M,P,S | 2I5Y | 0.78 |  |
VLM | VALINYLAMINE | G,M,P,S | 1YYL | 0.78 |  |
VLM | VALINYLAMINE | G,M,P,S | 2I60 | 0.78 |  |
LYN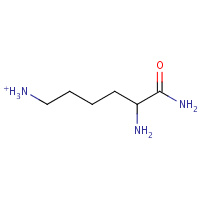 | 2,6-DIAMINO-HEXANOIC ACID AMIDE | A | 1GEA | 0.97 |  |
LYW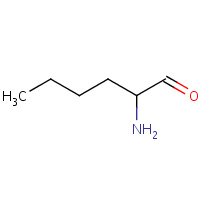 | 1-AMINO-1-CARBONYL PENTANE | A | 1EAG | 0.78 |  |
LPD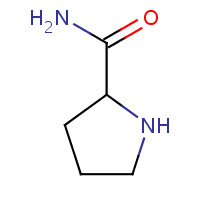 | L-PROLINAMIDE | H,S | 2H9E | 0.72 |  |
HPN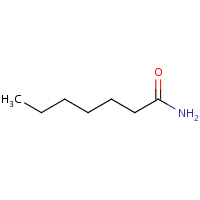 | HEPTANAMIDE | A,B | 1NWW | 0.72 |  |
NLN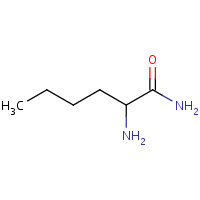 | NORLEUCINE AMIDE | A | 1DW6 | 0.97 |  |
NLN | NORLEUCINE AMIDE | A,B,C | 2AOE | 0.97 |  |
NLN | NORLEUCINE AMIDE | A,B | 1EBK | 0.97 |  |
RIN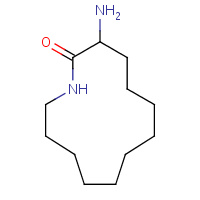 | 3-AMINO-AZACYCLOTRIDECAN-2-ONE | A,B | 1KBC | 0.73 |  |
LNO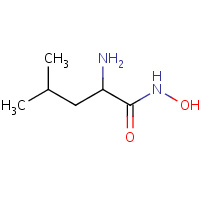 | L-LEUCYL-HYDROXYLAMINE | A | 4TLN | 0.74 |  |


