Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs03079465
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
MHL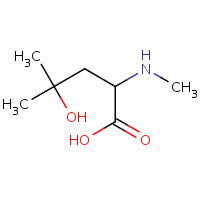 | N-METHYL-4-HYDROXY-LEUCINE | C | 1CWL | 0.73 |  |
DNM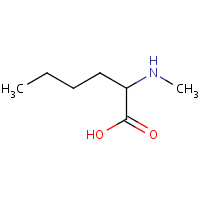 | N-METHYL-D-NORLEUCINE | A,B | 1S1O | 0.72 |  |
DNM | N-METHYL-D-NORLEUCINE | A,B | 1S4A | 0.72 |  |
DNM | N-METHYL-D-NORLEUCINE | A | 1R9V | 0.72 |  |
1PI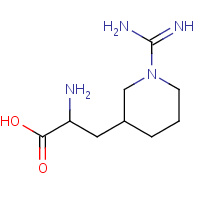 | 3-(1-CARBAMIMIDOYL-PIPERIDIN-3- YL)-L-ALANINE | A,B | 1ZZZ | 0.7 |  |
MCP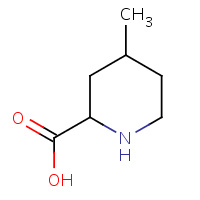 | 2-CARBOXY-4-METHYLPIPERIDINE | H | 1ETR | 0.86 |  |
LP6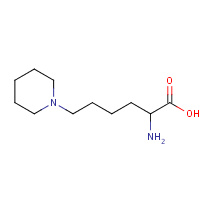 | 6-piperidin-1-yl-L-norleucine | A,B | 2W7Z | 0.71 |  |
MLE | N-METHYLLEUCINE | A,D | 2J9A | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1IKF | 0.72 |  |
MLE | N-METHYLLEUCINE | A,M,N | 2Z6W | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P | 1C5F | 0.72 |  |
MLE | N-METHYLLEUCINE | D,E,F | 1XQ7 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWA | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWF | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P,R,T | 2RMB | 0.72 |  |
MLE | N-METHYLLEUCINE | B | 3CYS | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWH | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,C,D,E,F, G,H | 3BO7 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CYN | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1LV9 | 0.72 |  |
MLE | N-METHYLLEUCINE | C,D | 2OJU | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P,R,T | 2RMA | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1IKM | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWK | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B | 2HG8 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWB | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWJ | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CYB | 0.72 |  |
MLE | N-METHYLLEUCINE | D | 1QNG | 0.72 |  |
MLE | N-METHYLLEUCINE | B | 1MIK | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CSA | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWL | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CYA | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,D | 1MF8 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1BCK | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,D,E,F,H | 1M63 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWC | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWI | 0.72 |  |
MLE | N-METHYLLEUCINE | C,D | 1QNH | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H | 2RMC | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWM | 0.72 |  |
MLE | N-METHYLLEUCINE | T,U,V | 2POY | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,C,D,E,F, I,J,K,L,M,N | 2ESL | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWO | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1IKL | 0.72 |  |
POM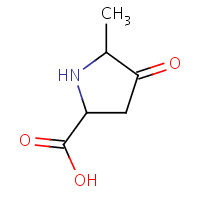 | CIS-5-METHYL-4-OXOPROLINE | A,B | 1A7Z | 0.74 |  |
NSK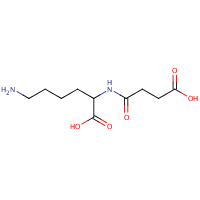 | N-SUCCINYL LYSINE | A | 2P8B | 0.7 |  |
CYJ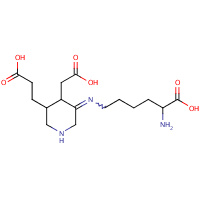 | (Z)-N~6~-[(4R,5S)-5-(2-CARBOXYETHYL)- 4-(CARBOXYMETHYL)PIPERIDIN-3-YLIDENE]- L-LYSINE | A,B | 2C14 | 0.73 |  |
XCP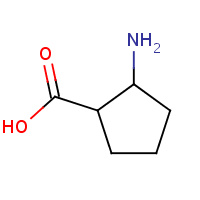 | (1S,2S)-2-aminocyclopentanecarboxylic acid | A | 3HF0 | 0.7 |  |
XCP | (1S,2S)-2-aminocyclopentanecarboxylic acid | A,B,D,E,F | 3FDM | 0.7 |  |
XCP | (1S,2S)-2-aminocyclopentanecarboxylic acid | A | 3C3H | 0.7 |  |
OIC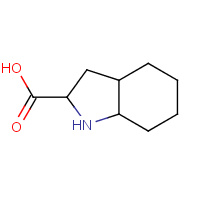 | OCTAHYDROINDOLE-2-CARBOXYLIC ACID | B,C | 3BV9 | 0.72 |  |
OIC | OCTAHYDROINDOLE-2-CARBOXYLIC ACID | A | 1BDK | 0.72 |  |
3GA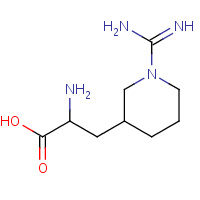 | 3-PIPERIDYL-N-GUANIDINO-L-ALANINE | B,C | 1BB0 | 0.7 |  |
3GA | 3-PIPERIDYL-N-GUANIDINO-L-ALANINE | 1 | 1YYY | 0.7 |  |
3GA | 3-PIPERIDYL-N-GUANIDINO-L-ALANINE | B,C | 1CA8 | 0.7 |  |
MNL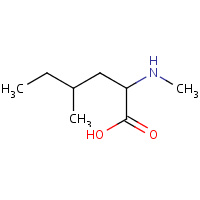 | 4,N-DIMETHYLNORLEUCINE | C | 1CWC | 0.77 |  |
DPL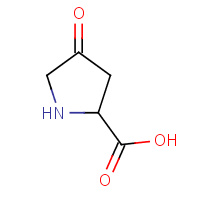 | 4-OXOPROLINE | A,B,C | 1QFI | 0.71 |  |
CPI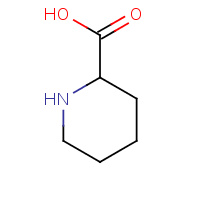 | 6-CARBOXYPIPERIDINE | A,B,C,D,F,Q | 1EFR | 0.79 |  |
CPI | 6-CARBOXYPIPERIDINE | B | 1EOL | 0.79 |  |
CPI | 6-CARBOXYPIPERIDINE | B | 1EOJ | 0.79 |  |
CPI | 6-CARBOXYPIPERIDINE | A,B,C,D,E,F, G,H,I,J,K,L | 1W3M | 0.79 |  |
CPI | 6-CARBOXYPIPERIDINE | A,B | 2ITK | 0.79 |  |
CPI | 6-CARBOXYPIPERIDINE | A,B | 2Q5A | 0.79 |  |
MCL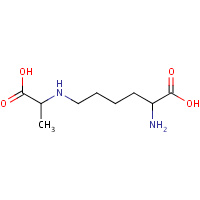 | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 1O5K | 0.72 |  |
MCL | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 3CPR | 0.72 |  |
MCL | NZ-(1-CARBOXYETHYL)-LYSINE | A,B | 2VC6 | 0.72 |  |
SSC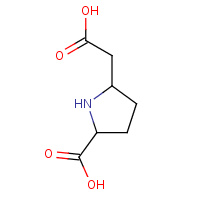 | (2S,5S)-5-CARBOXYMETHYLPROLINE | A,B,C,D | 1Q19 | 0.74 |  |
DPR | D-PROLINE | P | 1MCB | 0.7 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5B | 0.7 |  |
DPR | D-PROLINE | A | 2KDQ | 0.7 |  |
DPR | D-PROLINE | P | 1MCS | 0.7 |  |
DPR | D-PROLINE | A,B | 2AXI | 0.7 |  |
DPR | D-PROLINE | A | 1JY9 | 0.7 |  |
DPR | D-PROLINE | A | 2JUE | 0.7 |  |
DPR | D-PROLINE | P | 1MCK | 0.7 |  |
DPR | D-PROLINE | P | 1MCR | 0.7 |  |
DPR | D-PROLINE | A | 2D55 | 0.7 |  |
DPR | D-PROLINE | A | 1T8J | 0.7 |  |
DPR | D-PROLINE | A,B | 2EJ6 | 0.7 |  |
DPR | D-PROLINE | P | 1MCE | 0.7 |  |
DPR | D-PROLINE | P | 1MCI | 0.7 |  |
DPR | D-PROLINE | A,B | 1SNE | 0.7 |  |
DPR | D-PROLINE | A,B,C | 1QFI | 0.7 |  |
DPR | D-PROLINE | A,P | 1MCD | 0.7 |  |
DPR | D-PROLINE | A | 1ICL | 0.7 |  |
DPR | D-PROLINE | P | 1MCQ | 0.7 |  |
DPR | D-PROLINE | G,M,P,S | 2I5Y | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 3BOG | 0.7 |  |
DPR | D-PROLINE | A,L | 1ZEA | 0.7 |  |
DPR | D-PROLINE | A,B | 173D | 0.7 |  |
DPR | D-PROLINE | A,P | 1MCJ | 0.7 |  |
DPR | D-PROLINE | A,B,C | 1A7Y | 0.7 |  |
DPR | D-PROLINE | A | 1IC9 | 0.7 |  |
DPR | D-PROLINE | A,B | 1JY6 | 0.7 |  |
DPR | D-PROLINE | A | 209D | 0.7 |  |
DPR | D-PROLINE | A | 1BFW | 0.7 |  |
DPR | D-PROLINE | A,B | 1O6I | 0.7 |  |
DPR | D-PROLINE | A,B,C,D,E,F, G,H,I,J,K,L, M,N,O,P,Q,R, S,T,U,V,W,X | 1UNJ | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 2R3C | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1I3W | 0.7 |  |
DPR | D-PROLINE | A | 316D | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1SN9 | 0.7 |  |
DPR | D-PROLINE | A,B,C,H,K,L | 2R5D | 0.7 |  |
DPR | D-PROLINE | A,B | 2Q33 | 0.7 |  |
DPR | D-PROLINE | G,M,P,S | 2I60 | 0.7 |  |
DPR | D-PROLINE | A,B | 1JY4 | 0.7 |  |
DPR | D-PROLINE | P | 1MCF | 0.7 |  |
DPR | D-PROLINE | A,B,C,D,E,F | 1UNM | 0.7 |  |
DPR | D-PROLINE | A | 1ICO | 0.7 |  |
DPR | D-PROLINE | A,B | 1XOF | 0.7 |  |
DPR | D-PROLINE | P | 1MCC | 0.7 |  |
DPR | D-PROLINE | A,B,C,D | 1SNA | 0.7 |  |
X7O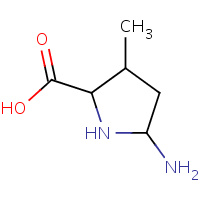 | 5-AMINO-3-METHYL-PYRROLIDINE-2- CARBOXYLIC ACID | A | 1L2Q | 0.71 |  |
2CG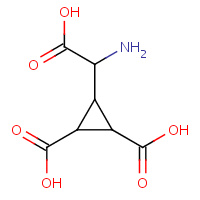 | (1R,2R)-3-[(S)-amino(carboxy)methyl]cyclopropane- 1,2-dicarboxylic acid | A,B | 2E4V | 0.71 |  |
IML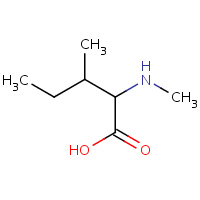 | N-METHYL-ISOLEUCINE | C | 1CWM | 0.72 |  |


