Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02968286
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
SVY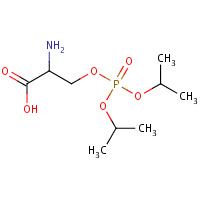 | O-[BIS(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A,B | 2QM0 | 0.76 |  |
SVY | O-[BIS(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A,B | 2JGI | 0.76 |  |
SGB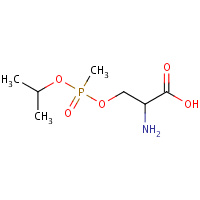 | O-[(S)-METHYL(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A | 3DT8 | 0.73 |  |
SGB | O-[(S)-METHYL(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A,B | 2WHP | 0.73 |  |
SGB | O-[(S)-METHYL(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A,B | 3F96 | 0.73 |  |
SGB | O-[(S)-METHYL(1-METHYLETHOXY)PHOSPHORYL]- L-SERINE | A,B | 2JGG | 0.73 |  |
SXE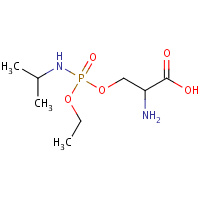 | O-{(S)-ETHOXY[(1-METHYLETHYL)AMINO]PHOSPHORYL}- L-SERINE | A,B | 2JGF | 0.73 |  |
SDP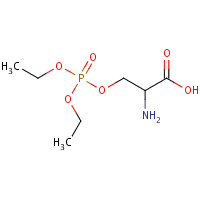 | 2-AMINO-3-(DIETHOXY-PHOSPHORYLOXY)- PROPIONIC ACID | A | 1J00 | 0.7 |  |
SDP | 2-AMINO-3-(DIETHOXY-PHOSPHORYLOXY)- PROPIONIC ACID | A | 3C6B | 0.7 |  |
MIS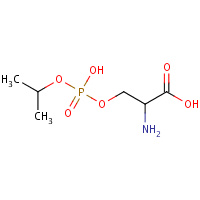 | MONOISOPROPYLPHOSPHORYLSERINE | A | 2DFP | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 1GNV | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 3GCN | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 3GCO | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A,B | 2JGM | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 2Z2X | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 1PPZ | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A,B,C | 3GDV | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 5PTP | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 3GDS | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A | 1A2Q | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A,B,C,D,E,F, G,H,I | 2RCE | 0.72 |  |
MIS | MONOISOPROPYLPHOSPHORYLSERINE | A,B,C | 3GDU | 0.72 |  |
LSP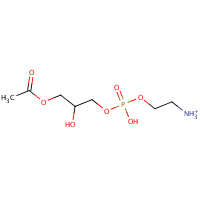 | (7S)-4,7-DIHYDROXY-10-OXO-3,5,9- TRIOXA-4-PHOSPHAUNDECAN-1-AMINIUM 4- OXIDE | A | 2DDE | 0.72 |  |


