Ligands with its PDB code and its tanimoto calculated from the molecule, present in the database, which has for MMscode: MMs02308041
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
You can sort the columns by Ligand, Ligand name, PDB and Tanimoto.
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
|---|---|---|---|---|---|
| Ligand | Ligand name | Chain | PDB | Tanimoto | Ligand in PDB |
GAM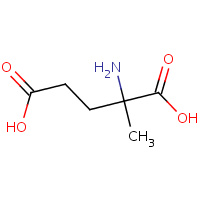 | ALPHA-METHYL-L-GLUTAMIC ACID | A,B | 1BJO | 0.71 |  |
DNM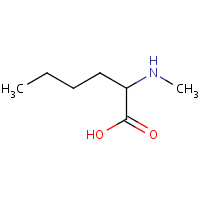 | N-METHYL-D-NORLEUCINE | A,B | 1S1O | 0.72 |  |
DNM | N-METHYL-D-NORLEUCINE | A,B | 1S4A | 0.72 |  |
DNM | N-METHYL-D-NORLEUCINE | A | 1R9V | 0.72 |  |
RGP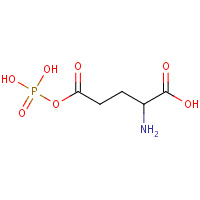 | GAMMA-GLUTAMYL PHOSPHATE | A,B | 2J5V | 0.71 |  |
SN0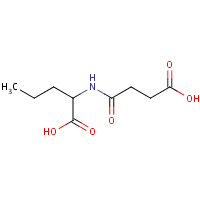 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | A | 2G65 | 0.73 |  |
SN0 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | A | 2G6A | 0.73 |  |
SN0 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | A | 2G68 | 0.73 |  |
SN0 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | C,D,E,X,Y,Z | 2FG7 | 0.73 |  |
SN0 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | A | 2G6C | 0.73 |  |
SN0 | N-(3-CARBOXYPROPANOYL)-L-NORVALINE | C,D,E,X,Y,Z | 2FG6 | 0.73 |  |
DM0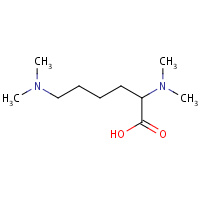 | N~2~,N~2~,N~6~,N~6~-tetramethyl- L-lysine | A | 132L | 0.77 |  |
MLE | N-METHYLLEUCINE | A,D | 2J9A | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1IKF | 0.72 |  |
MLE | N-METHYLLEUCINE | A,M,N | 2Z6W | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P | 1C5F | 0.72 |  |
MLE | N-METHYLLEUCINE | D,E,F | 1XQ7 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWA | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWF | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P,R,T | 2RMB | 0.72 |  |
MLE | N-METHYLLEUCINE | B | 3CYS | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWH | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,C,D,E,F, G,H | 3BO7 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CYN | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1LV9 | 0.72 |  |
MLE | N-METHYLLEUCINE | C,D | 2OJU | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H,J,L, N,P,R,T | 2RMA | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1IKM | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWK | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B | 2HG8 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWB | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWJ | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CYB | 0.72 |  |
MLE | N-METHYLLEUCINE | D | 1QNG | 0.72 |  |
MLE | N-METHYLLEUCINE | B | 1MIK | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CSA | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWL | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1CYA | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,D | 1MF8 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1BCK | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,D,E,F,H | 1M63 | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWC | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWI | 0.72 |  |
MLE | N-METHYLLEUCINE | C,D | 1QNH | 0.72 |  |
MLE | N-METHYLLEUCINE | B,D,F,H | 2RMC | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWM | 0.72 |  |
MLE | N-METHYLLEUCINE | T,U,V | 2POY | 0.72 |  |
MLE | N-METHYLLEUCINE | A,B,C,D,E,F, I,J,K,L,M,N | 2ESL | 0.72 |  |
MLE | N-METHYLLEUCINE | C | 1CWO | 0.72 |  |
MLE | N-METHYLLEUCINE | A | 1IKL | 0.72 |  |
AN0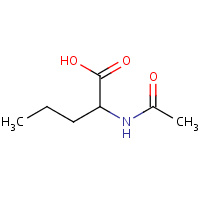 | N-ACETYL-L-NORVALINE | C,D,E,X,Y,Z | 2G7M | 0.71 |  |
AN0 | N-ACETYL-L-NORVALINE | A | 1ZQ8 | 0.71 |  |
SYM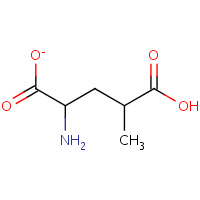 | 2S,4R-4-METHYLGLUTAMATE | A,B | 1SD3 | 0.75 |  |
NIG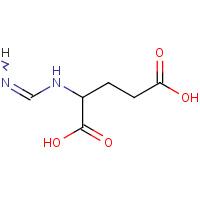 | N-(IMINOMETHYL)-L-GLUTAMIC ACID | A,B | 2PUZ | 0.76 |  |
NLQ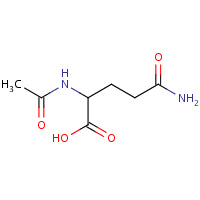 | N~2~-ACETYL-L-GLUTAMINE | A,B,C,D | 2GGH | 0.73 |  |
NLQ | N~2~-ACETYL-L-GLUTAMINE | A,B,C,D | 1XPY | 0.73 |  |
MEG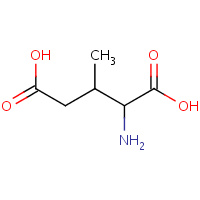 | (2S,3R)-3-METHYL-GLUTAMIC ACID | A | 1T5N | 0.75 |  |
MEG | (2S,3R)-3-METHYL-GLUTAMIC ACID | A | 1T5M | 0.75 |  |
NLG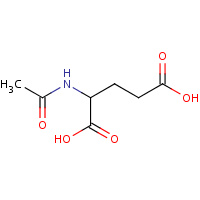 | N-ACETYL-L-GLUTAMATE | A | 1OH9 | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A | 1OHA | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A,B,C,D,E,F, G,H,I,J,K,L | 2BUF | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A,B,C,D | 2RD5 | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A,B | 2JJ4 | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A,B,C,D,E,F, G,H,J,K | 2V5H | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A | 1GSJ | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A | 3B8G | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A | 1GS5 | 0.81 |  |
NLG | N-ACETYL-L-GLUTAMATE | A,B,C | 2BTY | 0.81 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFX | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C,H,K,L | 2R5D | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1C4B | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1CW8 | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,D | 1CZQ | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFY | 0.79 |  |
DGL | D-GLUTAMIC ACID | I | 1QUR | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,D | 2Q3I | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1CVQ | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 1P4N | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2VVT | 0.79 |  |
DGL | D-GLUTAMIC ACID | E,S | 148L | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1KR6 | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C,D | 2GZM | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1AY3 | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 2J8F | 0.79 |  |
DGL | D-GLUTAMIC ACID | U | 2AIZ | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C | 2DWU | 0.79 |  |
DGL | D-GLUTAMIC ACID | L | 2EAX | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFZ | 0.79 |  |
DGL | D-GLUTAMIC ACID | H,I | 4THN | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1CWZ | 0.79 |  |
DGL | D-GLUTAMIC ACID | P | 1MCK | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 2JUE | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFO | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 3H41 | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 1D0K | 0.79 |  |
DGL | D-GLUTAMIC ACID | L,N | 1WCO | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C | 1ZUW | 0.79 |  |
DGL | D-GLUTAMIC ACID | H,I | 5GDS | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C,H,K,L | 2R5B | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFQ | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2Q33 | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B | 2JFP | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,E,F | 2W4I | 0.79 |  |
DGL | D-GLUTAMIC ACID | A | 2J8G | 0.79 |  |
DGL | D-GLUTAMIC ACID | A,B,C,D | 2R3C | 0.79 |  |
SHR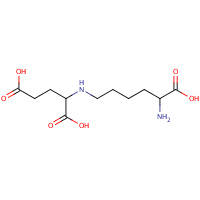 | N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID | A,B,C,D,E,F, G,H | 1E5Q | 0.76 |  |
N7P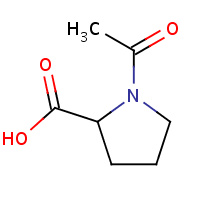 | 1-ACETYL-D-PROLINE | A,B,C | 1NX8 | 0.7 |  |
LAL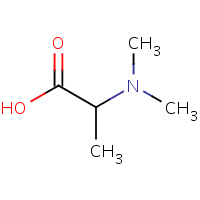 | N,N-DIMETHYL-L-ALANINE | A,B | 1R1G | 0.73 |  |
IML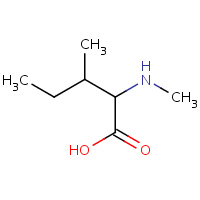 | N-METHYL-ISOLEUCINE | C | 1CWM | 0.72 |  |
PBE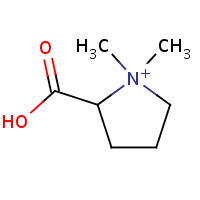 | 1,1-DIMETHYL-PROLINIUM | A,B | 2B4M | 0.75 |  |
PBE | 1,1-DIMETHYL-PROLINIUM | A | 1R9Q | 0.75 |  |
PBE | 1,1-DIMETHYL-PROLINIUM | A,B | 1SW1 | 0.75 |  |
LME | (3R)-3-METHYL-L-GLUTAMIC ACID | A | 1XT7 | 0.75 |  |


